Seasonality and Immunity
Note: This blogpost was conceptualised in mid-2021, and should be public late in 2023. With the problems going on at 𝕏 (formerly Twitter), it's struck me that putting any such content on a website that I control probably has its advantages, so I hope to use this site more going forwards.
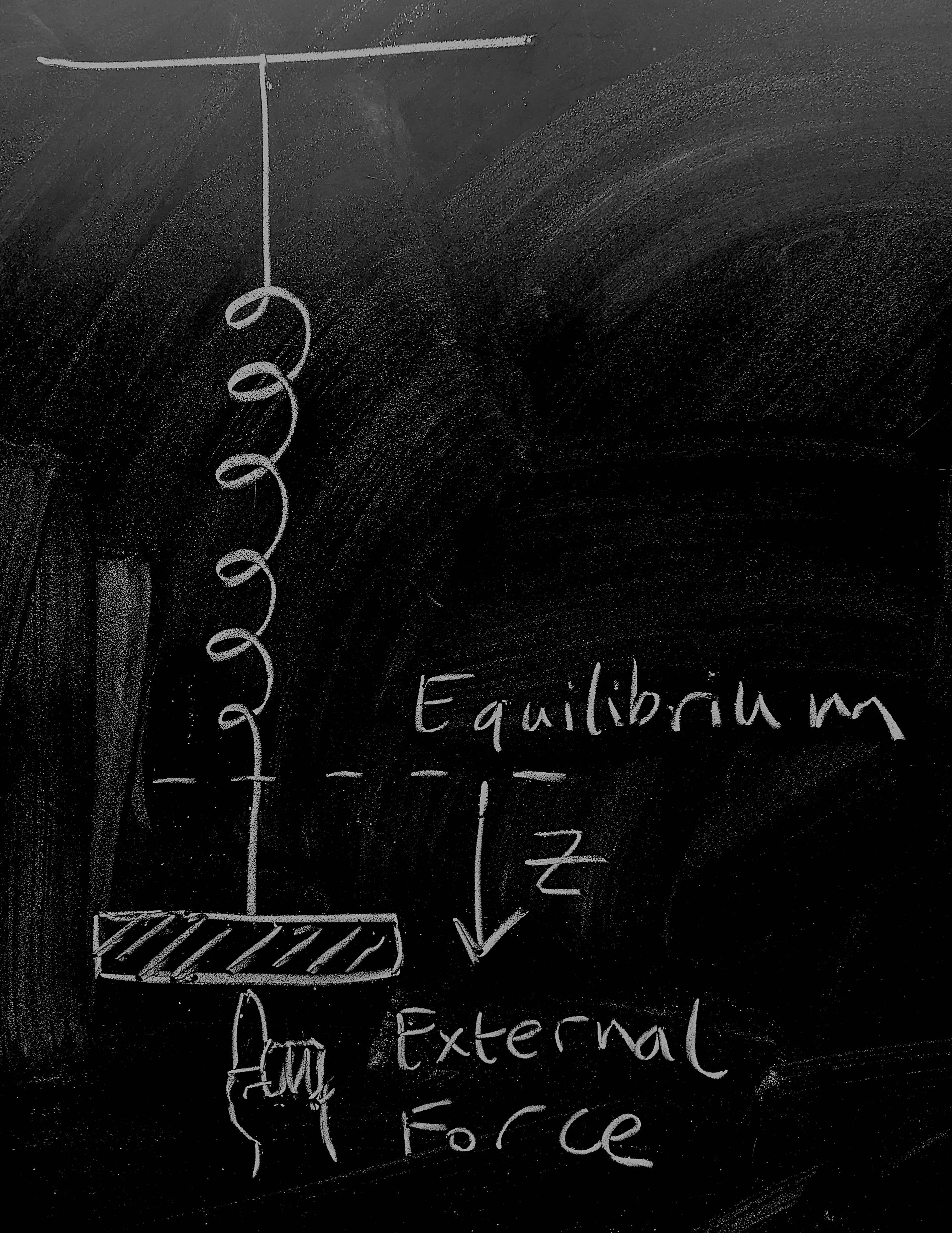
So let's start by considering a weight on a spring as pictured - this is going to give us a physical 'model' of disease dynamics for an endemic infection. The weight on the spring is a distance \(z(t)\) from the point at which the system would be at rest. There are three forces acting on the weight:
The spring pulls it towards equilibrium, to a first approximation, proportionally to its spring constant giving a force contribution \(-k z\);
As the weight moves, friction dissipates kinetic energy giving a force contribution \(-\nu \dot{z}\);
A human is pushing at the weight giving a time-dependent force contribution \(\varphi(t)\).
Now, dredging up our high-school physics we remember that Newton's equation is \(F = ma\). Noting that acceleration is the second derivative of position so that \(a = \ddot{z}\) and combining the three force contributions above we get \[-kz - \nu \dot{z} + \varphi(t) = m \ddot{z}\, .\] Dredging still further, we might remember that momentum is \(p = m\dot{z}\), which gives us dynamics \[\dot{z} = \frac{1}{m} p \, , \qquad \dot{p} = -kz - \frac{\nu}{m} p + \varphi(t) \, . \qquad \mathrm{(1)} \]
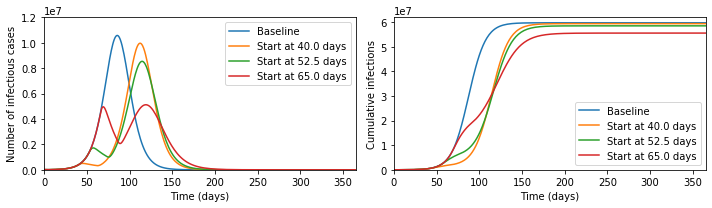
Next, let us consider an SIRS model with forcing, \[\dot{S} = \omega R - \beta(t) S I \, , \quad\ \dot{I} = \beta(t) S I - \gamma I \, , \quad \dot{R} = \gamma I - \omega R \, .\] This models a system where individuals move from susceptible to infections upon contact with other infectious individuals, then recover, and after that immunity wanes and they become susceptible again. The rate \(\beta(t)\) depending on time is supposed to capture different exogenous events: schools opening and closing; new vaccines; new variants; policy changes; and all sorts of other complexity. If you want to experiment with these equations numerically, check out this Jupyter notebook that I make available on GitHub.
This set of equations does not have a general closed form, but we can look for an approximate one using the Ansatz \[S(t) = S_* + \epsilon x(t) \, , \quad\ I(t) = I_* + \epsilon y(t) \, , \quad R(t) = N - S(t) - I(t) \, , \quad \beta(t) = \beta_* + \epsilon \phi(t) \, .\] We want this to correspond to a fixed point of the dynamical system when \(\epsilon = 0\), leading to the equations \[0 = \omega (N - S_* - I_*) - \beta_* S_* I_* \, , \qquad\ 0 = \beta_* S_* I_* - \gamma I_* \, .\] These have a solution when \(S_* = N, I_* = 0\) that is unstable if the disease can go endemic, i.e. when \(\mathcal{R}_0 = \beta_*/\gamma > 1 \); the other fixed point is when \(S_* = 1/\mathcal{R}_0\), which I hope to blog about later. The expression for the expected number of infectives \(I_*\) at endemicity is more complex; feel free to derive it for fun! For our purposes here, we are interested in what we obtain at first order in \(\epsilon\), which gives us \[\dot{x} = - \omega x - \omega y - \beta_* x I_* - \beta_* S_* y - \phi S_* I_* + \mathcal{O}(\epsilon^2) \, , \qquad\ \dot{y} = \beta_* x I_* + \beta_* S_* y + \phi S_* I_* - \gamma y + \mathcal{O}(\epsilon^2) \, . \qquad \mathrm{(2)} \]
OK, so take a breath. Now, with a bit of work, we can write both equations (1) and (2) (neglecting quadratic terms in \(\epsilon\)) above in the form of a two-dimensional vector differential equation \[\dot{\mathbf{v}} = \boldsymbol{J} \mathbf{v} + \mathbf{F}(t) \, , \qquad \mathrm{(3)} \] where \(\mathbf{v}\) is a vector containing the dynamical variables - \((z,p)\) for the spring and \((x,y)\) for the SIRS model - \(\boldsymbol{J}\) is a matrix depending on rate constants, and \(\mathbf{F}(t)\) is a vector function of time. Some readers will note that the details of the versions of (3) derived from (1) and (2) are not quite the same, but also that equation (3) takes the same structure under the linear transformation \[\tilde{\mathbf{v}} = \boldsymbol{T} \mathbf{v} \, ,\quad \tilde{\boldsymbol{J}} = \boldsymbol{T}\boldsymbol{J}\boldsymbol{T}^{-1} \, ,\quad \tilde{\mathbf{F}} = \boldsymbol{T} \mathbf{F} \, . \] They might also have fun trying to work with (3) via matrix integrating factor, Eigensystem analysis, or maybe Fourier transform.
So, that was quite fiddly (although not much beyond high-school level mathematics apart from the last bit). But the TL;DR is this: imagine all the complexity that you can generate by bouncing a weight up and down. If your taps line up just right with the how the spring behaves, you can get regular oscillations, but it's not difficult to make the weight jump irregularly all over the place. And this is exactly what we can see with an endemic disease: there's no guarantee that oscillatory behaviour will be regular or predictable.
Now, for many diseases that are better established, we do indeed see relatively regular annual peaks, although predicting the exact timing and height of these even for very well studied diseases like influenza is not currently possible. But most experts think that the relative regularity of influenza compared to COVID arises because the exogenous forces are mainly annual events like school holidays, people moving in and outdoors with the weather, and maybe even a direct bio-physical impact of the weather, with evolutionary events generally having a less dramatic impact on disease transmission than they do for COVID.
Because evolution is hard to predict, it's not clear that COVID will become more like influenza. Or even that it would be a good thing if it did. The purpose of this post, however, was to help explain why its unpredictability is at least somewhat predictable.