This worksheet is based on the tenth lecture of the Statistical Modelling in Stata course, created by Dr Mark Lunt and offered by the Centre for Epidemiology Versus Arthritis at the University of Manchester. The original Stata exercises and solutions are here translated into their R equivalents.
Refer to the original slides and Stata worksheets here.
Life tables and survival curves
This section uses the dataset leukaemia.csv.
leukaemia <- read.csv('leukaemia.csv')
First, load the survival package.
The time variable is weeks, the number of weeks to relapse. The outcome variable is relapse, which is 1 if the subject had a relapse at that time and 0 if they did not. Create a corresponding survival object using the syntax Surv(time, status). This will be on the left hand side of your model formula.
First reorder the levels of wbc3cat so that ‘Normal’ is the reference group (for ease of comparison with the Stata solutions sheet). Similarly we can relevel treatment1 so that ‘Standard’ is the first level (as otherwise ‘Drug A’ comes first, alphabetically).
Obtain a life table for the subjects on Drug A. What is the median survival in this group (at what time does the survival function reach 0.5)?
survA <- survfit(Surv(weeks, relapse) ~ 1,
data = leukaemia,
subset = treatment1 == 'Drug A')
summary(survA, times = unique(leukaemia$weeks)) # `times` is optional
Call: survfit(formula = Surv(weeks, relapse) ~ 1, data = leukaemia,
subset = treatment1 == "Drug A")
time n.risk n.event survival std.err lower 95% CI upper 95% CI
1 21 0 1.000 0.0000 1.000 1.000
2 21 0 1.000 0.0000 1.000 1.000
3 21 0 1.000 0.0000 1.000 1.000
4 21 0 1.000 0.0000 1.000 1.000
5 21 0 1.000 0.0000 1.000 1.000
6 21 3 0.857 0.0764 0.720 1.000
7 17 1 0.807 0.0869 0.653 0.996
8 16 0 0.807 0.0869 0.653 0.996
9 16 0 0.807 0.0869 0.653 0.996
10 15 1 0.753 0.0963 0.586 0.968
11 13 0 0.753 0.0963 0.586 0.968
12 12 0 0.753 0.0963 0.586 0.968
13 12 1 0.690 0.1068 0.510 0.935
15 11 0 0.690 0.1068 0.510 0.935
16 11 1 0.627 0.1141 0.439 0.896
17 10 0 0.627 0.1141 0.439 0.896
19 9 0 0.627 0.1141 0.439 0.896
20 8 0 0.627 0.1141 0.439 0.896
22 7 1 0.538 0.1282 0.337 0.858
23 6 1 0.448 0.1346 0.249 0.807
25 5 0 0.448 0.1346 0.249 0.807
32 4 0 0.448 0.1346 0.249 0.807
34 2 0 0.448 0.1346 0.249 0.807
35 1 0 0.448 0.1346 0.249 0.807At 23 weeks, the survival function drops below 0.5.
How many subjects were lost to followup in this treatment arm?
table(leukaemia$relapse, leukaemia$treatment1)
Standard Drug A
0 0 12
1 21 912 subjects were lost to followup on Drug A.
Obtain a life table for the subjects on standard treatment. What is the median survival in this group?
Call: survfit(formula = Surv(weeks, relapse) ~ 1, data = leukaemia,
subset = treatment1 == "Standard")
time n.risk n.event survival std.err lower 95% CI upper 95% CI
1 21 2 0.9048 0.0641 0.78754 1.000
2 19 2 0.8095 0.0857 0.65785 0.996
3 17 1 0.7619 0.0929 0.59988 0.968
4 16 2 0.6667 0.1029 0.49268 0.902
5 14 2 0.5714 0.1080 0.39455 0.828
8 12 4 0.3810 0.1060 0.22085 0.657
11 8 2 0.2857 0.0986 0.14529 0.562
12 6 2 0.1905 0.0857 0.07887 0.460
15 4 1 0.1429 0.0764 0.05011 0.407
17 3 1 0.0952 0.0641 0.02549 0.356
22 2 1 0.0476 0.0465 0.00703 0.322
23 1 1 0.0000 NaN NA NAThe survival function drops below 0.5 after 8 weeks.
How many subjects were lost to followup in this treatment arm?
Zero (see table output above).
Do the answers to your previous questions suggest that Drug A is better, worse, or the same as standard treatment?
Median survival before relapse is better on Drug A (23 weeks) than standard treatment (8 weeks).
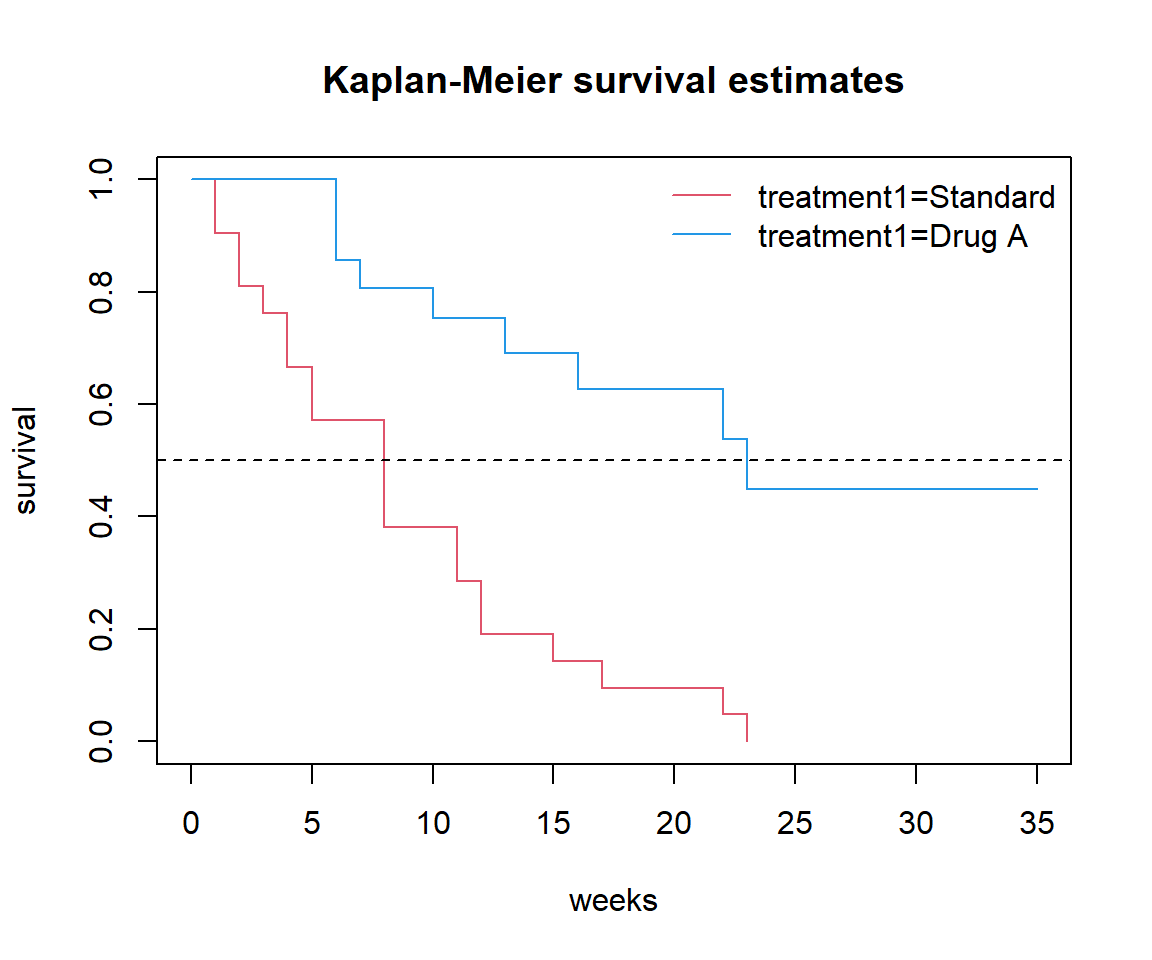
Produce a Kaplan-Meier curve for each of the treatments. Does this confirm your answer to the previous question?
There are some basic plotting functions in the survival package.
surv_t1 <- survfit(Surv(weeks, relapse) ~ treatment1, data = leukaemia)
plot(surv_t1, xlab = 'weeks', ylab = 'survival',
col = c(2, 4), main = 'Kaplan-Meier survival estimates')
legend('topright',
names(surv_t1$strata),
col = c(2, 4), lty = 1,
bty = 'n')
abline(h = 0.5, lty = 2) # median line
More tools for visualisation via ggplot2 are provided in the survminer package, for which a helpful cheat sheet is available. There’s less chance of mixing up the strata, as a legend is generated automatically by default:
library(survminer)
ggsurvplot(surv_t1, surv.median.line = 'hv')
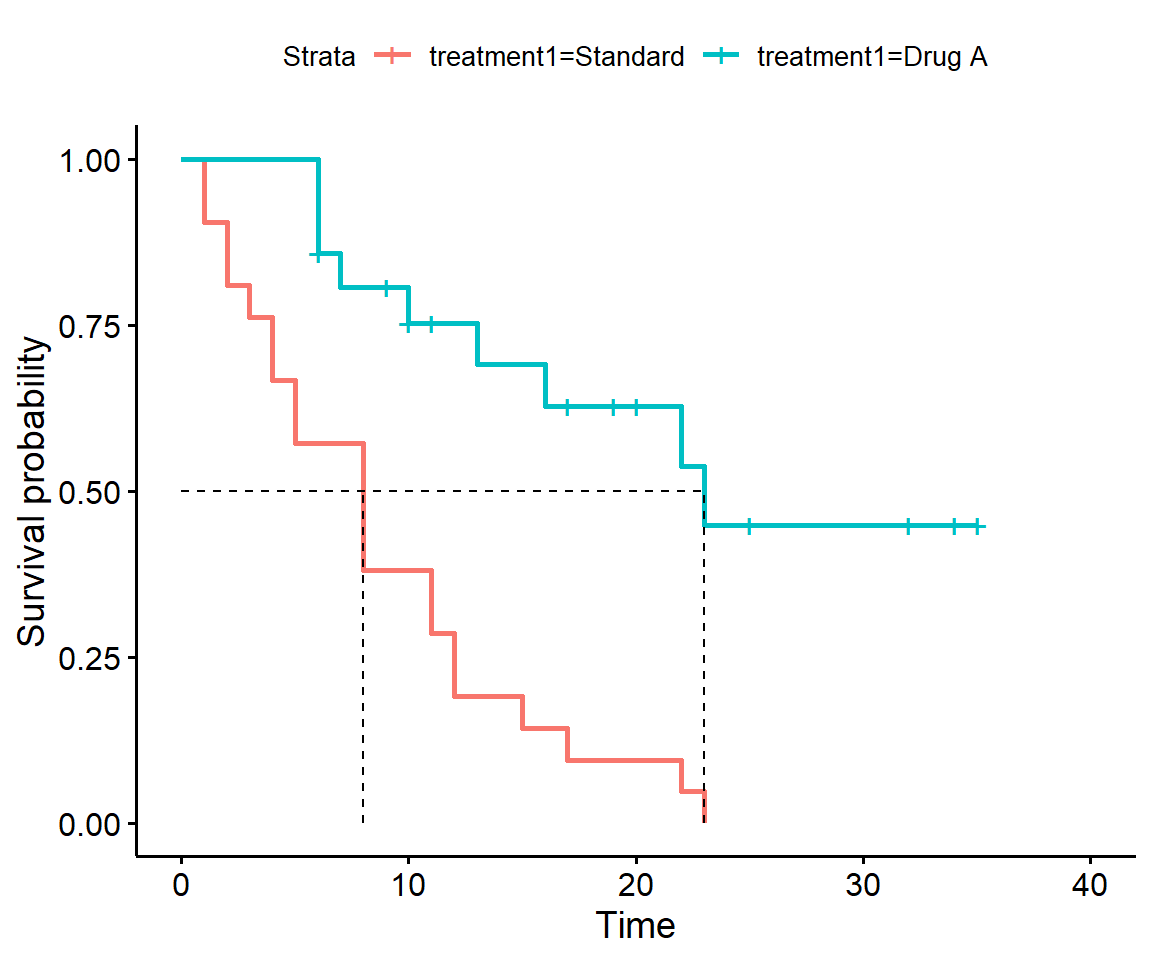
Add a horizontal line to the graph at y = 0.5. This line represents half of the group surviving and half having a relapse: the point where it crosses the two survival curves should give you the median survival times you calculated in earlier questions.
In base R graphics you can use abline and in ggplot2 there is geom_abline or geom_hline. The ggsurvplot function has a dedicated argument called surv.median.line for this purpose. See above.
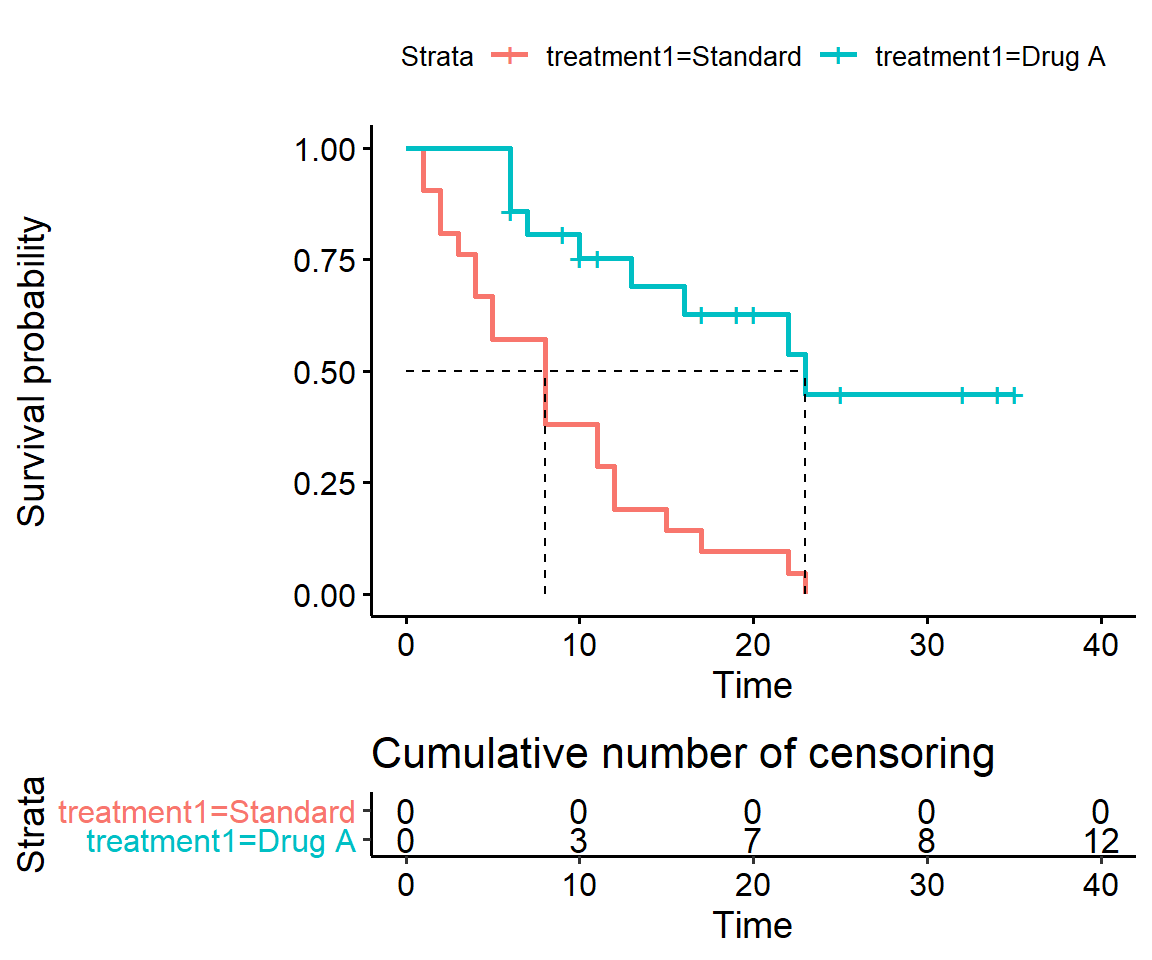
Add the number of subjects lost to followup in the two treatment arms to the corresponding time points on the graph.
This is slightly contrived because R doesn’t have a direct equivalent to the lost command. So here we’ll show what’s offered in survival and survminer. See the survminer vignette for more tips.
ggsurvplot(surv_t1,
surv.median.line = 'hv',
censor = TRUE,
cumcensor = TRUE,
ncensor.plot = TRUE)
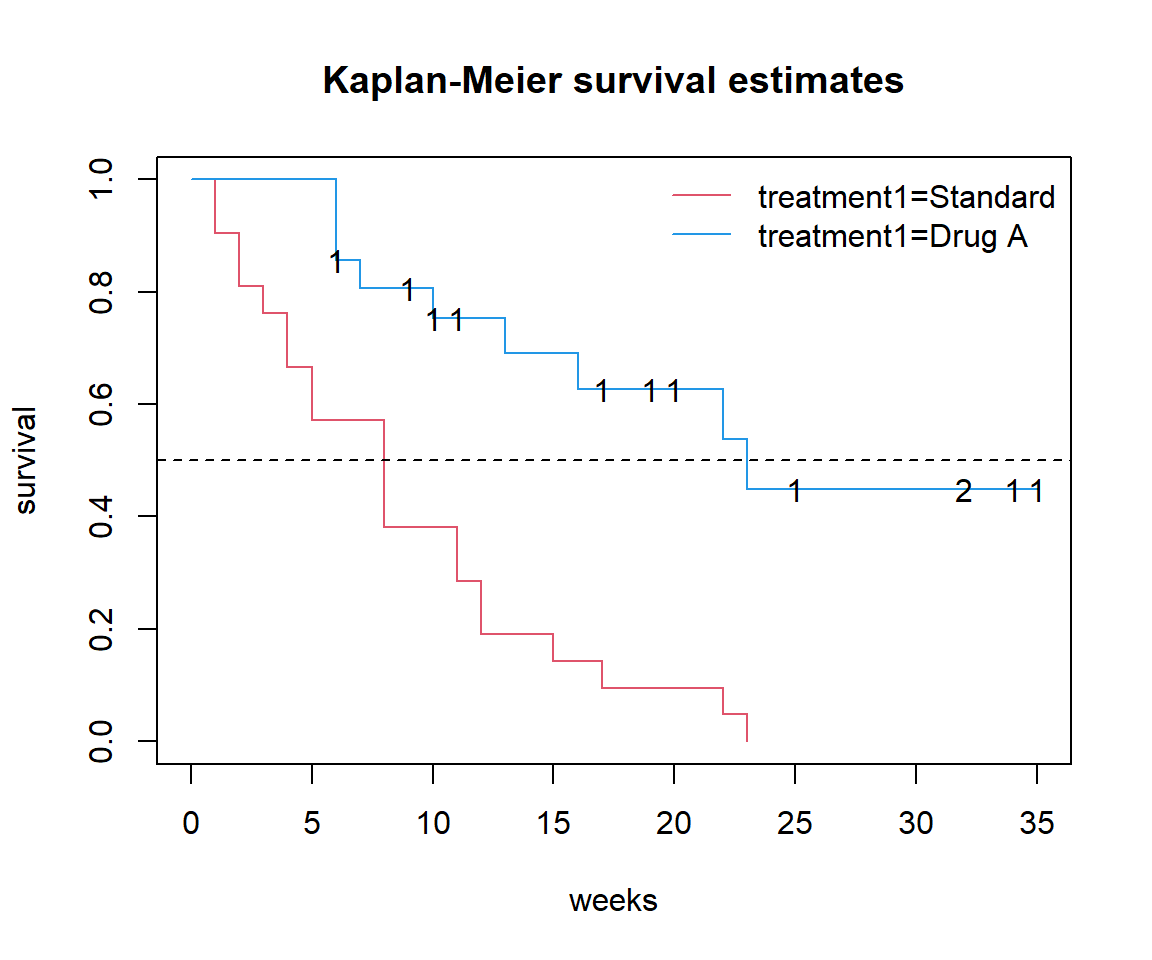
In base R, use the generic plotting functions text() or points() and extract the coordinates and values from the survfit object.
# Original plot:
plot(surv_t1, xlab = 'weeks', ylab = 'survival',
col = c(2, 4), main = 'Kaplan-Meier survival estimates')
legend('topright', names(surv_t1$strata),
col = c(2, 4), lty = 1, bty = 'n')
abline(h = 0.5, lty = 2)
# Add censor labels:
text(surv_t1$time, surv_t1$surv,
labels = surv_t1$n.censor,
# make label transparent if n.censor == 0
col = rgb(0, 0, 0, 0:1)[1 + (surv_t1$n.censor > 0)])
Add confidence bands to your plot(s).
Look in the documentation, depending on the plotting method you are using. Both base plot.survfit and survminer::ggsurv offer arguments conf.int = TRUE:
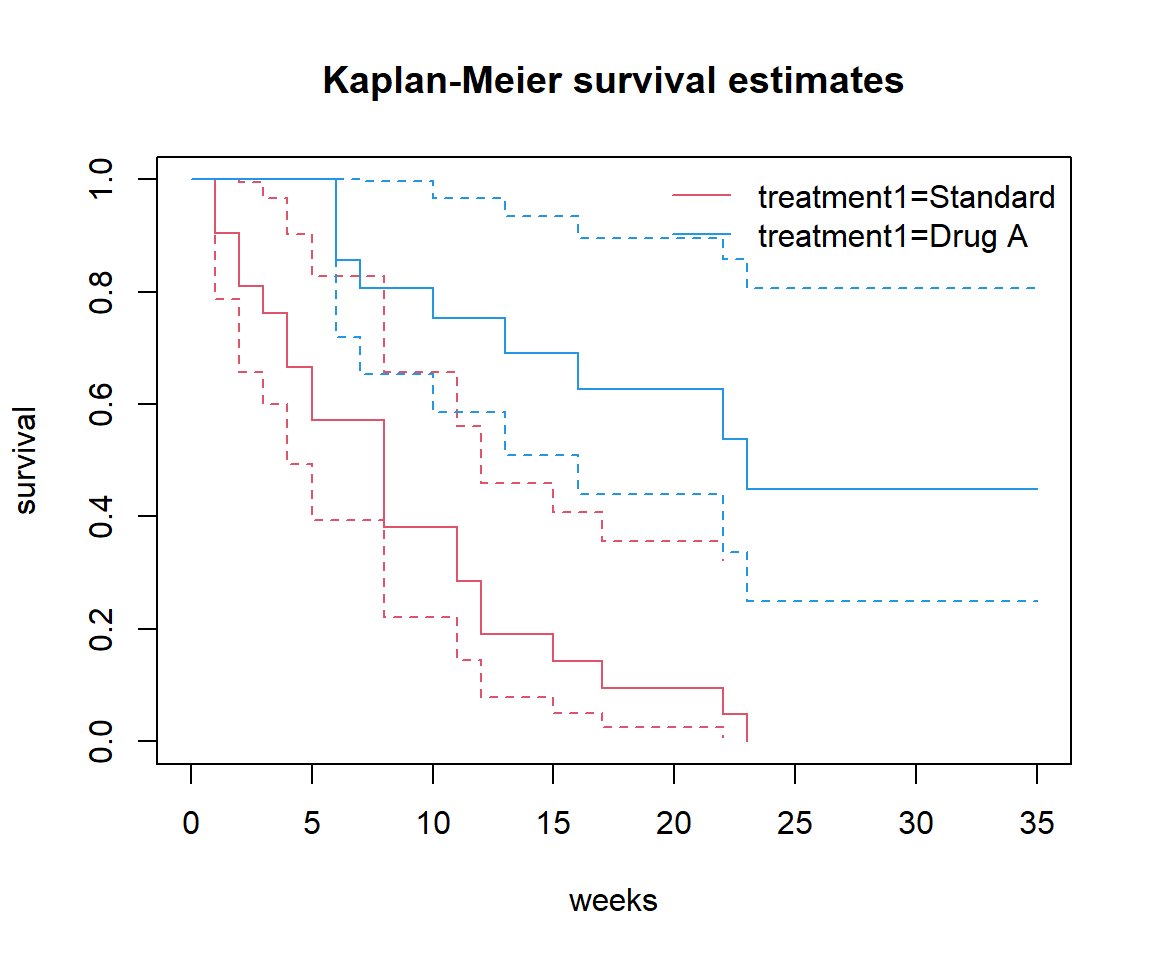
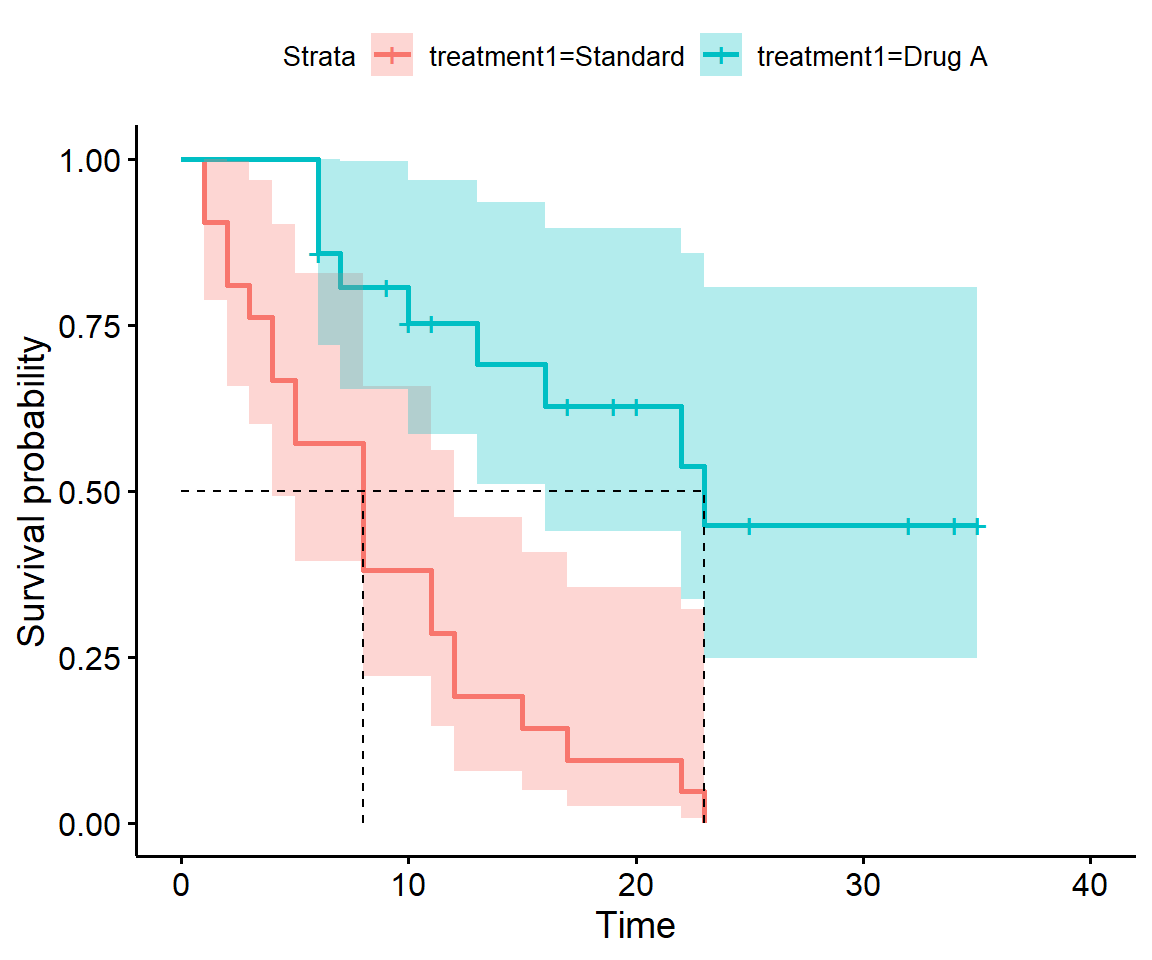
Why do the confidence bands get wider over time? Because they are based on smaller numbers.
Use survdiff to perform a logrank test to compare the survival on Drug A to that on standard treatment. Is the difference between Drug A and standard treatment statistically significant?
You can actually just do this test and add it to your survminer plot with the arguments pval and pval.method both set to TRUE:
ggsurvplot(surv_t1,
conf.int = TRUE,
censor = TRUE,
pval = TRUE, pval.method = TRUE)
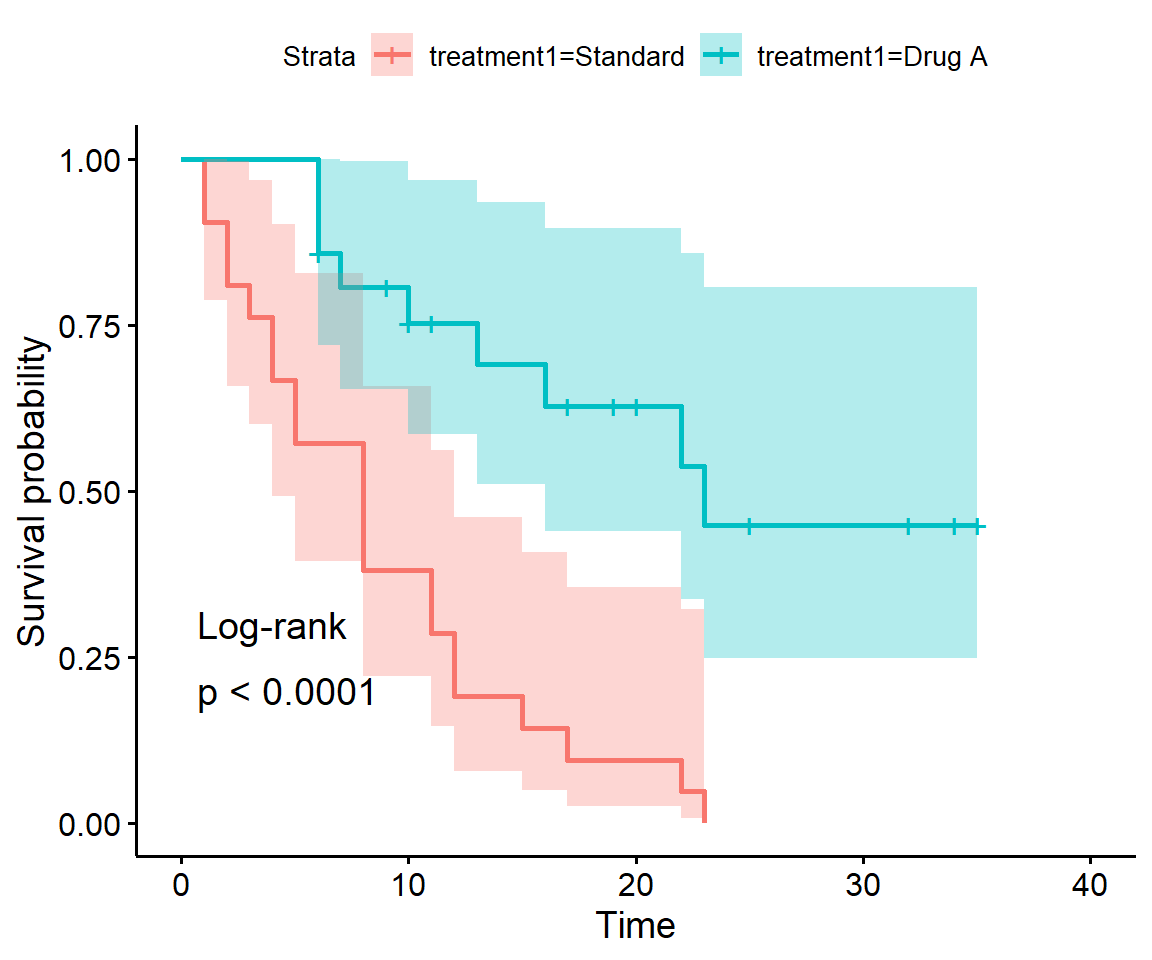
Or do it the old-fashioned way using survdiff():
Call:
survdiff(formula = Surv(weeks, relapse) ~ treatment1, data = leukaemia)
N Observed Expected (O-E)^2/E (O-E)^2/V
treatment1=Standard 21 21 10.7 9.77 16.8
treatment1=Drug A 21 9 19.3 5.46 16.8
Chisq= 16.8 on 1 degrees of freedom, p= 4e-05 The difference is statistically significant: there are fewer relapses on Drug A than expected.
Would you have had the same answer to the previous question if you had used a Wilcoxon test in place of a logrank test?
Call:
survdiff(formula = Surv(weeks, relapse) ~ treatment1, data = leukaemia,
rho = 1)
N Observed Expected (O-E)^2/E (O-E)^2/V
treatment1=Standard 21 14.55 7.68 6.16 14.5
treatment1=Drug A 21 5.12 12.00 3.94 14.5
Chisq= 14.5 on 1 degrees of freedom, p= 1e-04 Cox regression
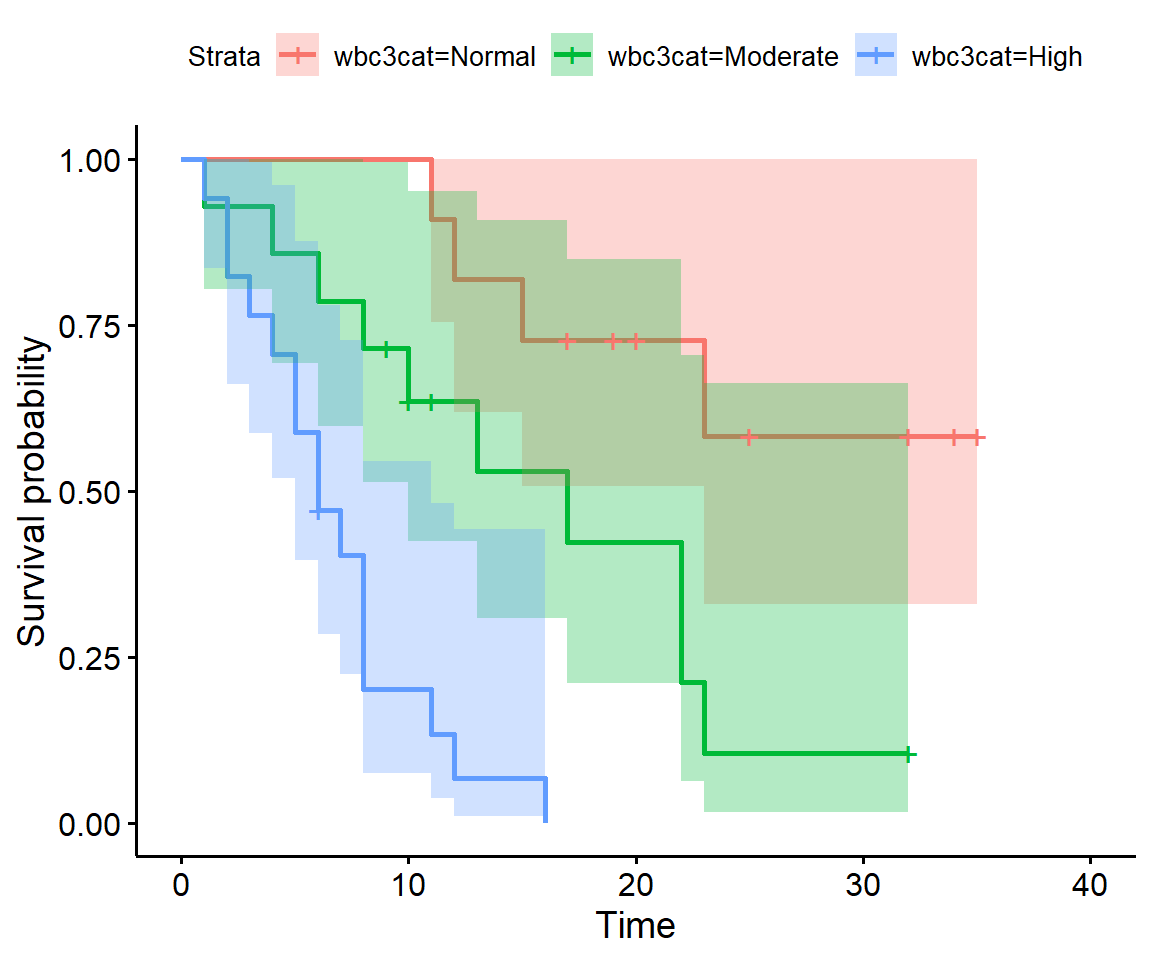
Have a look at the survival curves by white blood cell count. Does the white blood cell count affect survival?
surv_wbc <- survfit(Surv(weeks, relapse) ~ wbc3cat, data = leukaemia)
ggsurvplot(surv_wbc, conf.int = TRUE)
Do a cross-tabulation of treatment1 against wbc3cat. Are the proportions of subjects in each of the white blood cell counts categories the same in the two treatment arms?
library(magrittr)
xtabs(~ treatment1 + wbc3cat, data = leukaemia) %T>%
print %>%
proportions(margin = 1)
wbc3cat
treatment1 Normal Moderate High
Standard 4 5 12
Drug A 7 9 5 wbc3cat
treatment1 Normal Moderate High
Standard 0.1904762 0.2380952 0.5714286
Drug A 0.3333333 0.4285714 0.2380952The standard treatment appears to have a greater proportion of people with high white blood cell counts, compared to those on Drug A.
Tip. The %T>% pipe from magrittr lets us print and pass the result of xtabs to the next function, without having to save the table to a temporary variable.
Given that proportion of subjects in the “High” cell count group is greater in the standard treatment arm than in the Drug A arm, would you expect this to have increased or decreased survival in this arm of the trial?
Decreased, because leukaemia is a disease that causes high numbers of abnormal blood cells.
White blood cell count is a potential confounder, so we need to adjust for it. First, we will perform an unadjusted Cox regression to obtain the hazard ratio before adjusting. (Use function coxph().) What is the hazard ratio for Drug A, and its 95% confidence interval?
Call:
coxph(formula = Surv(weeks, relapse) ~ treatment1, data = leukaemia)
n= 42, number of events= 30
coef exp(coef) se(coef) z Pr(>|z|)
treatment1Drug A -1.5721 0.2076 0.4124 -3.812 0.000138 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
exp(coef) exp(-coef) lower .95 upper .95
treatment1Drug A 0.2076 4.817 0.09251 0.4659
Concordance= 0.69 (se = 0.041 )
Likelihood ratio test= 16.35 on 1 df, p=5e-05
Wald test = 14.53 on 1 df, p=1e-04
Score (logrank) test = 17.25 on 1 df, p=3e-05The hazard ratio and confidence interval are given by
2.5 % 97.5 %
treatment1Drug A 0.2076035 0.09251284 0.4658729Now obtain the adjusted hazard ratio. What is the adjusted hazard ratio and its 95% confidence interval?
Fit the adjusted model and extract the hazard ratio and its 95% confidence interval:
HR 2.5 % 97.5 %
0.2647984 0.1125064 0.6232375 How did the confounding by white blood cell count affect the apparent effect of Drug A? Is this what you expected from the earlier questions?
The beneficial effect of Drug A was exaggerated by the difference in white blood cell counts between the groups. The hazard ratio is larger (i.e. chance of relapse increased) after adjusting for it.
Now we need to test the proportional hazards assumption. First for treatment: produce a plot of the observed and predicted Kaplan Meier plots. Are the observed and predicted curves close to each other?
To check the proportional hazards assumption, we can use cox.zph() from the survival package or ggcoxzph() from the survminer package. Check the documentation also for information on ggcoxdiagnostics() and ggadjustedcurves().
For example:
We can also plot the observed versus expected survival curves, using survfit.
dummy <- list(treatment1 = levels(leukaemia$treatment1))
cox_surv <- survfit(cox_unadj, newdata = dummy)
plot(surv_t1, col = c(2, 4), lty = 1,
xlab = 'weeks', ylab = 'survival')
lines(cox_surv, col = c(2, 4), lty = 2)
legend('topright',
c('observed', 'expected', levels(leukaemia$treatment1)),
lty = c(1, 2, 4, 4), col = c(1, 1, 2, 4))
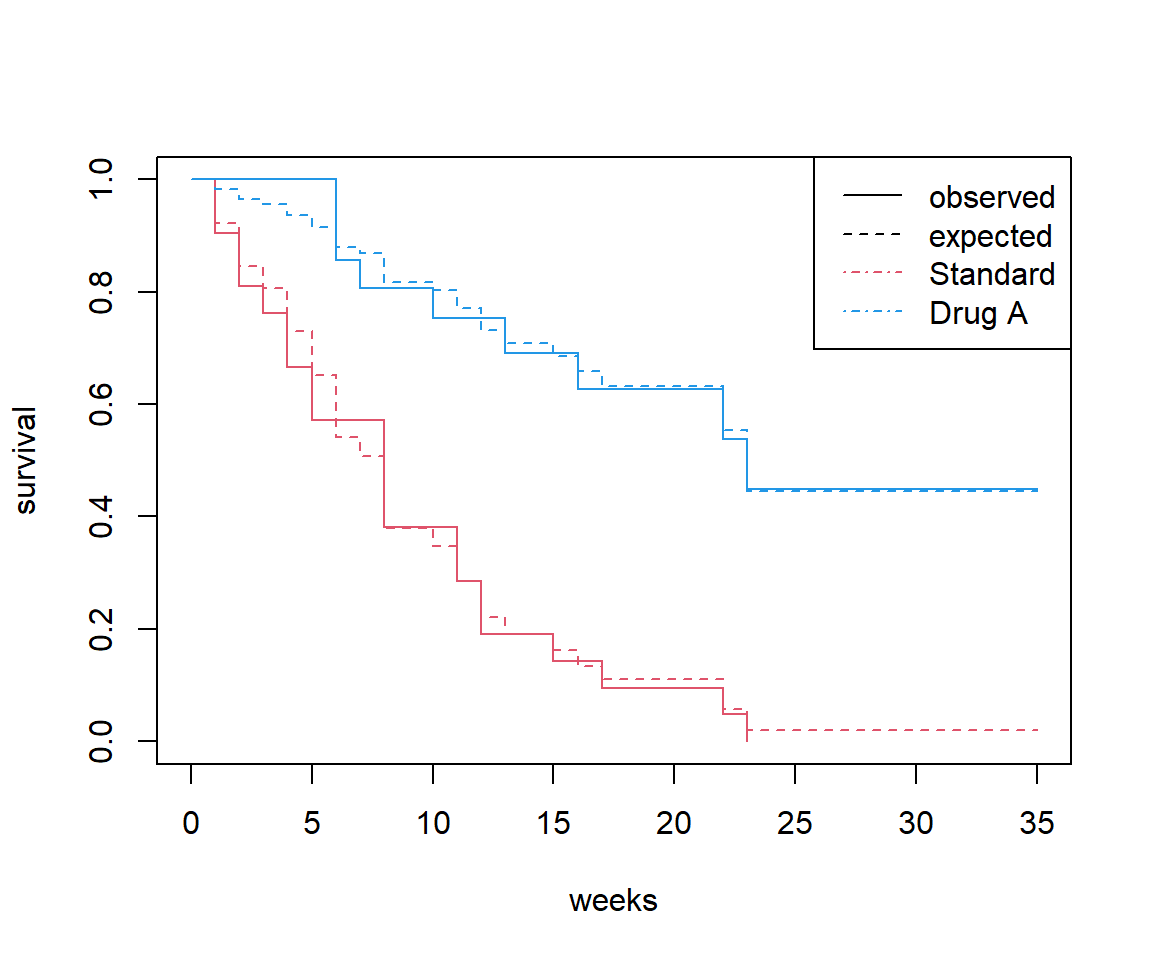
(If there is a ggplot2 or survminer approach, then it is left as an exercise.)
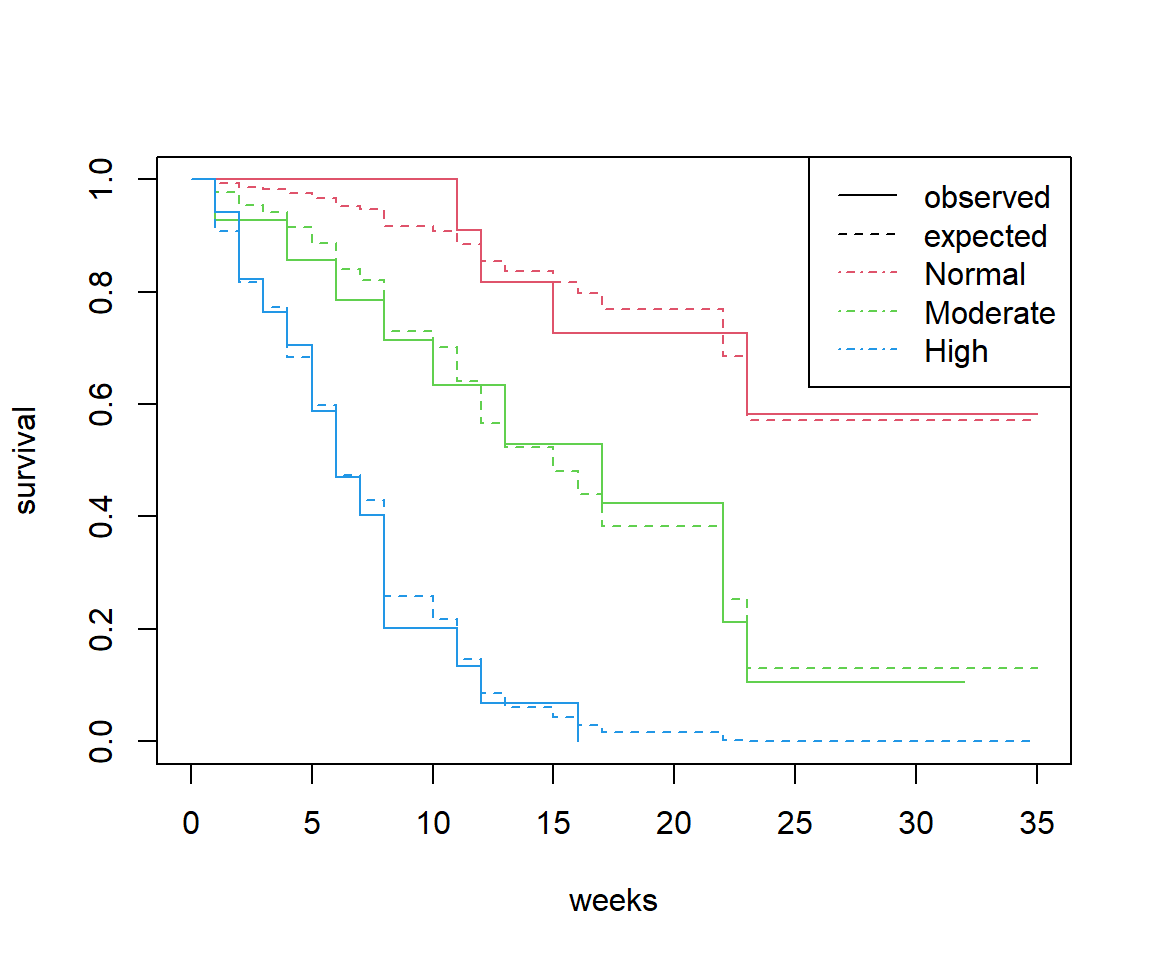
Now we can test the same assumption for the effect of white blood cell count. Are the observed and predicted curves close to each other?
dummy <- list(wbc3cat = levels(leukaemia$wbc3cat))
cox_surv <- survfit(cox_wbc, newdata = dummy)
plot(surv_wbc, col = c(2:4), lty = 1,
xlab = 'weeks', ylab = 'survival')
lines(cox_surv, col = c(2:4), lty = 2)
legend('topright',
c('observed', 'expected', levels(leukaemia$wbc3cat)),
lty = c(1, 2, 4, 4, 4), col = c(1, 1, 2:4))
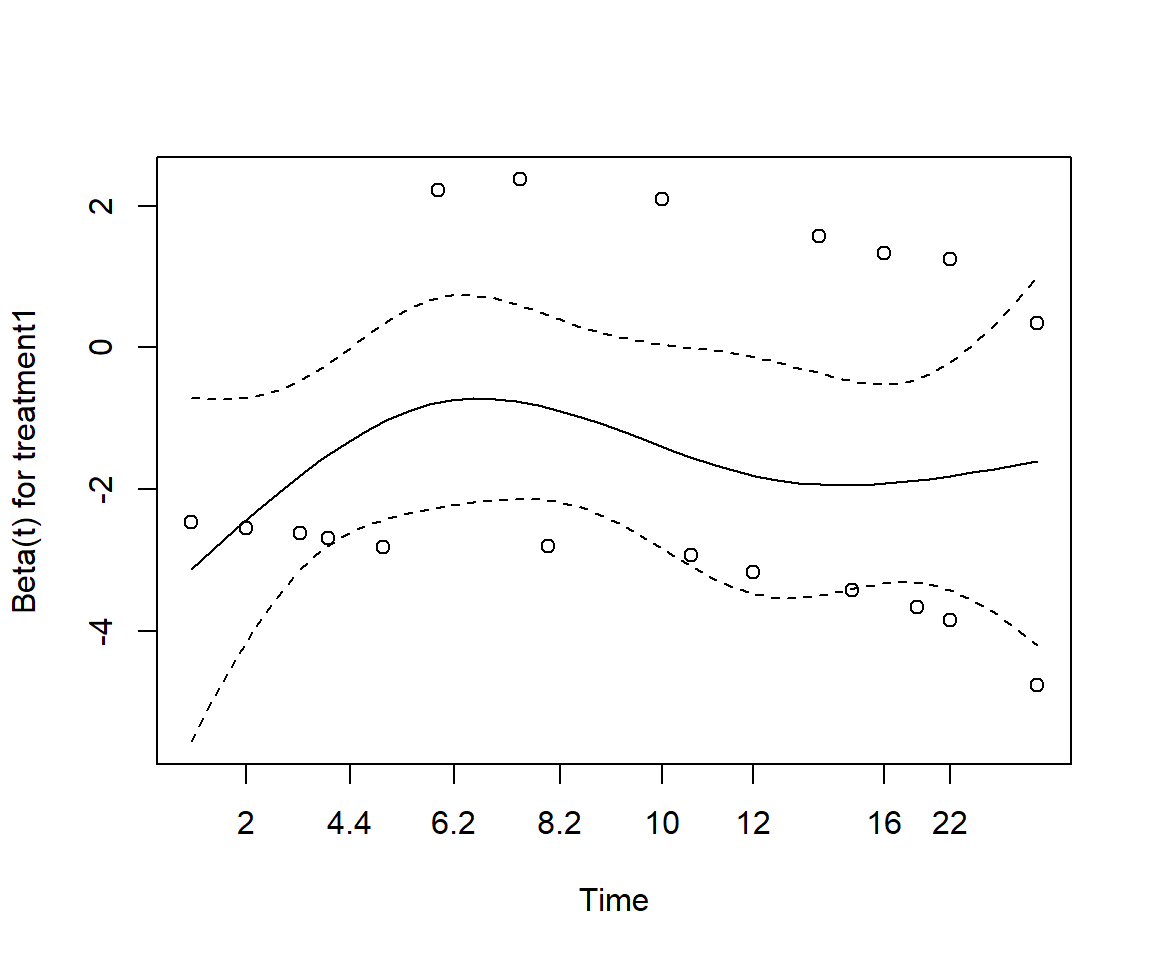
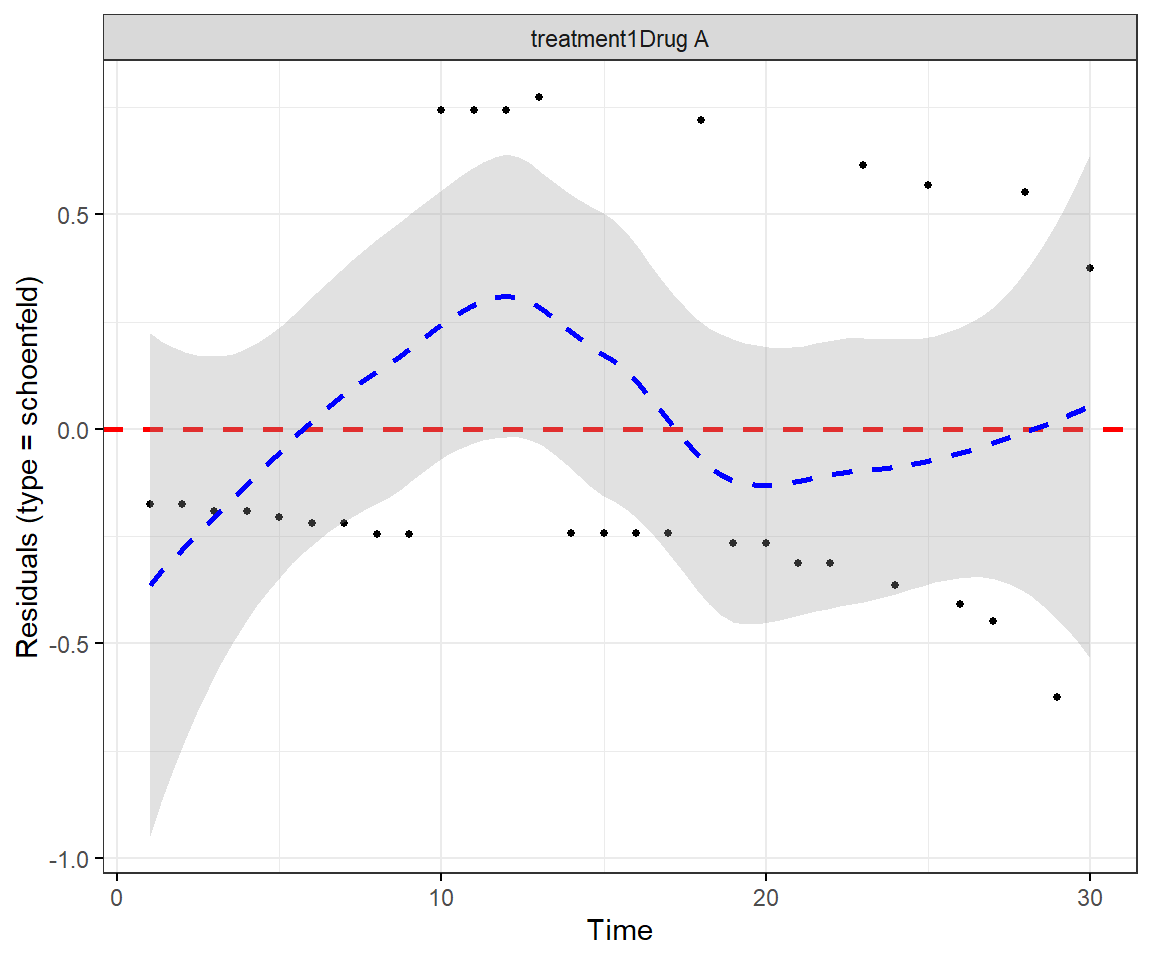
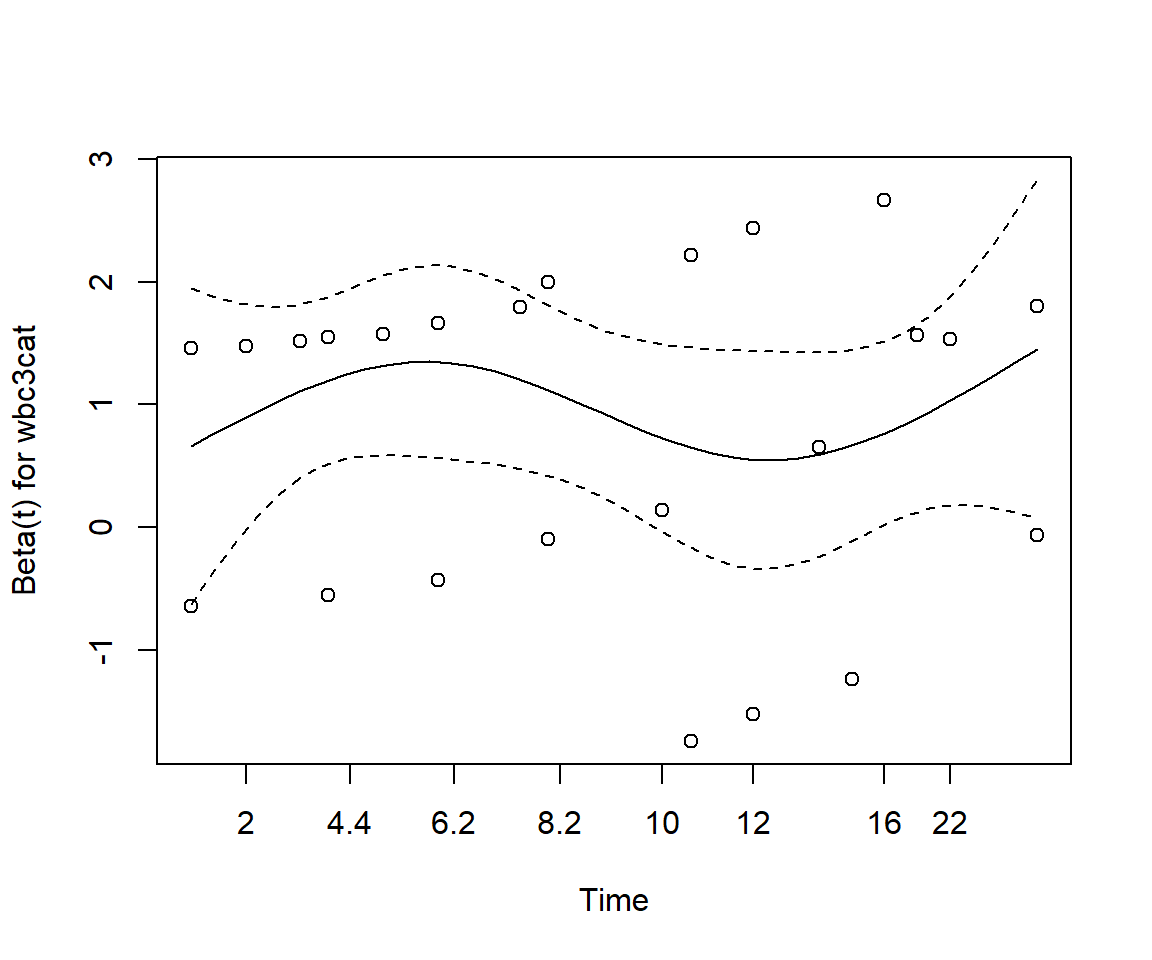
Perform a test of proportionality using Schoenfeld residuals. Is the regression model valid?
Note. You can regenerate both Schoenfeld residual plots at once with
To perform the test, simply print
cox.zph(cox_adj)
chisq df p
treatment1 0.181 1 0.67
wbc3cat 0.696 2 0.71
GLOBAL 0.813 3 0.85None of the test statistics is significant, thereby implying that the proportional hazards assumption holds for each of the variables.
Obtain tests of proportionality for each individual variable. Is there any evidence of non-proportional hazards?
The cox.zph() function did both sets of tests at once, so the answer is already given above.
Non-proportional hazards
There is a second drug used in this trial, stored in treatment2. Compare the survival curves for Drug B and standard treatment. How does the survival on Drug B compare to that on standard treatment during the first 10 weeks?
surv_t2 <- survfit(Surv(weeks, relapse) ~ treatment2, data = leukaemia)
plot(surv_t2, col = c(2, 4), mark.time = TRUE)
legend('topright', lty = 1, col = c(2, 4), names(surv_t2$strata))
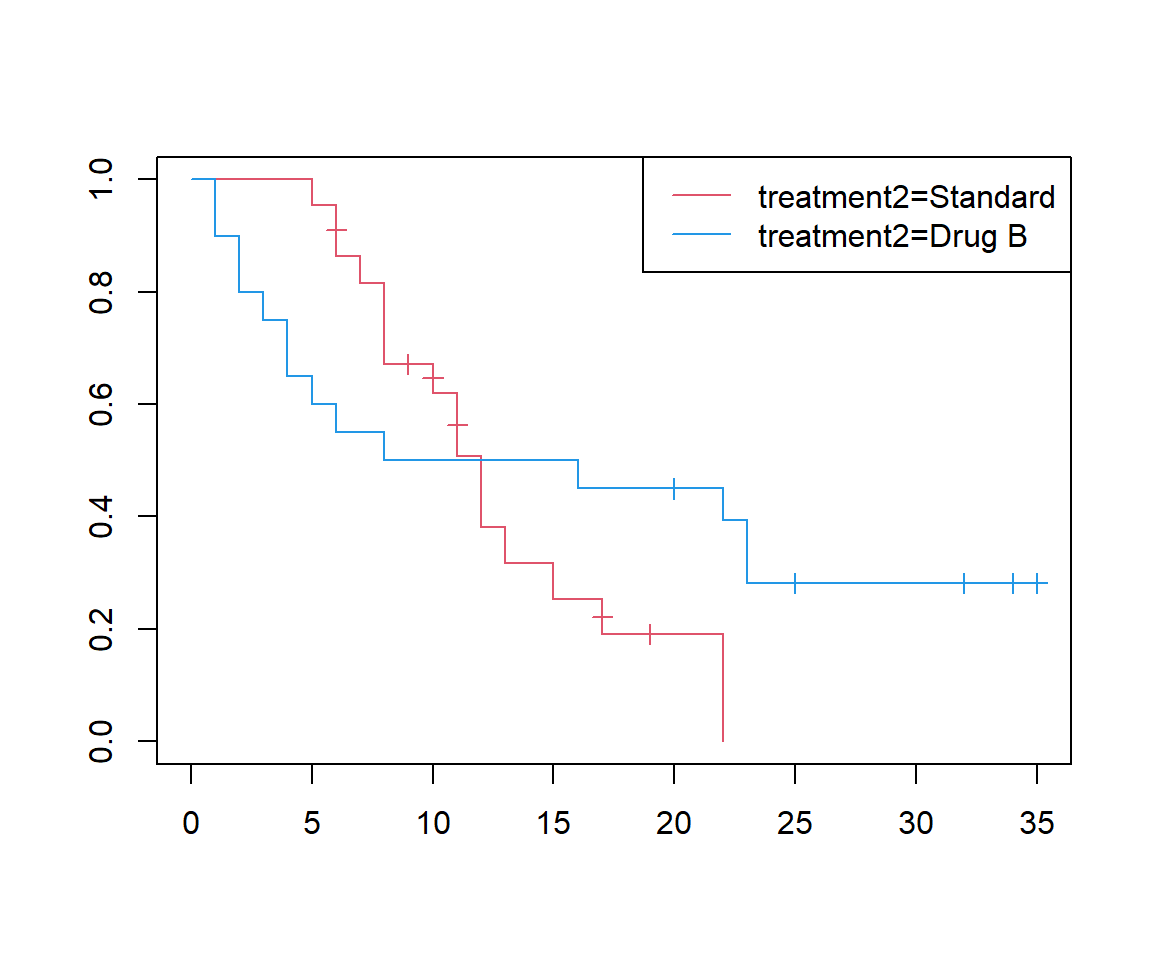
ggsurvplot(surv_t2)
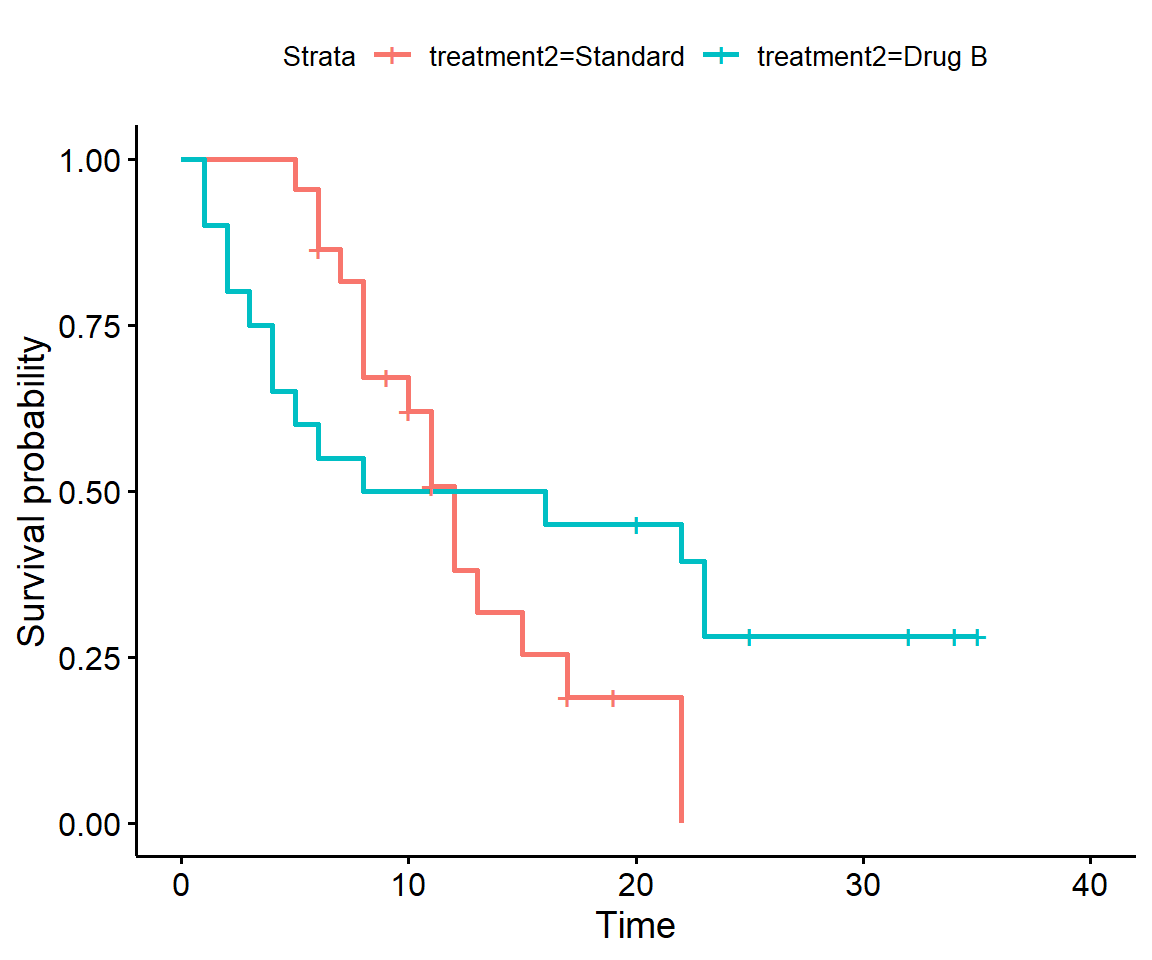
In the first 10 weeks, Drug B has worse survival than the standard treatment.
How does the survival on Drug B compare to that on standard treatment after the first 10 weeks?
After 10 weeks, survival on Drug B is better than the standard treatment.
Superimpose the predicted survival curves from a Cox regression model. How do the predicted and observed curves differ?
cox_t2 <- coxph(Surv(weeks, relapse) ~ treatment2, data = leukaemia)
dummy <- list(treatment2 = levels(leukaemia$treatment2))
cox_surv2 <- survfit(cox_t2, newdata = dummy)
plot(surv_t2, col = c(2, 4))
lines(cox_surv2, col = c(2, 4), lty = 4, lwd = 2)
legend('topright', lty = 1, col = c(2, 4), names(surv_t2$strata))
legend('bottomleft', lty = c(1, 4), lwd = 1:2, c('observed', 'predicted'))
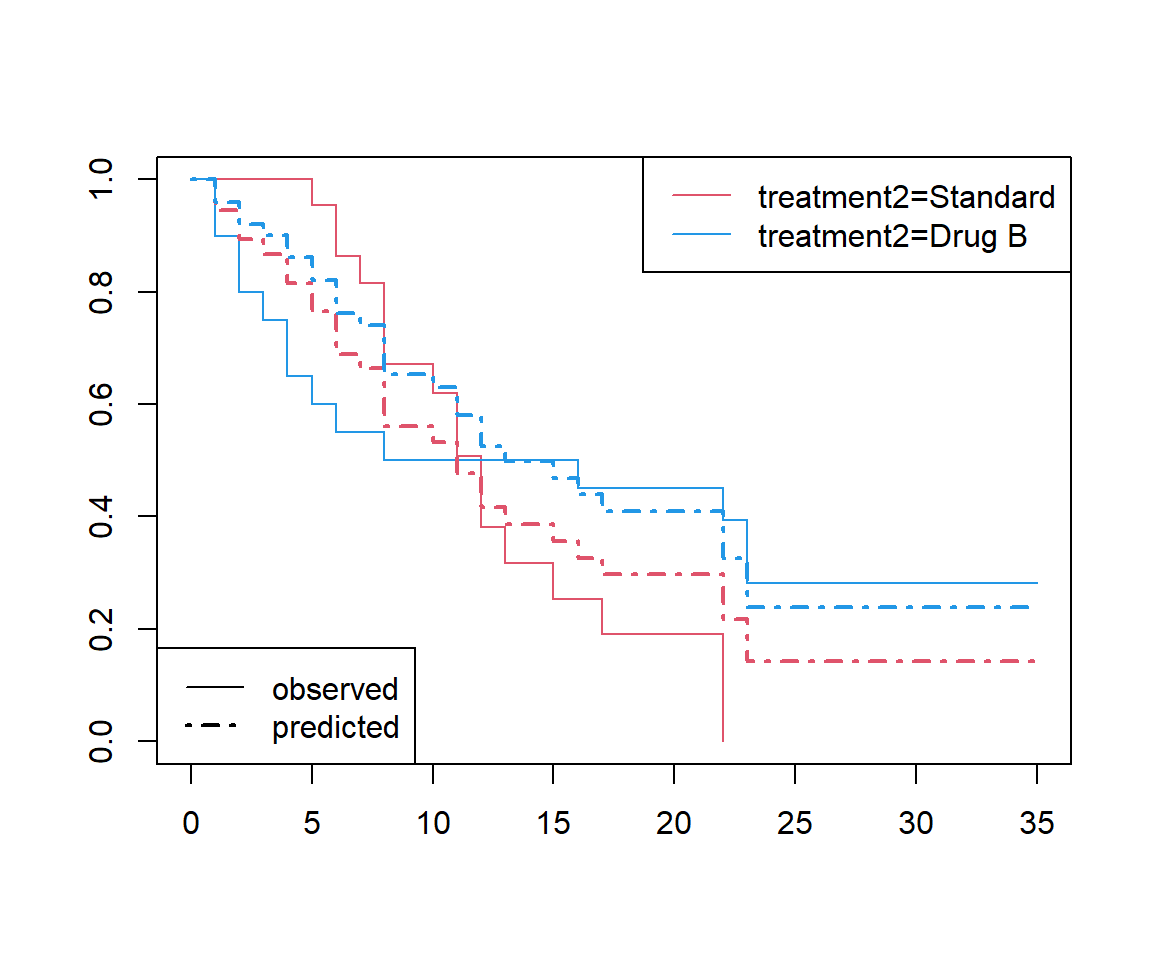
The observed survival curves intersect each other but the predicted curves do not (obviously, because they are based on a proportional hazards assumption).
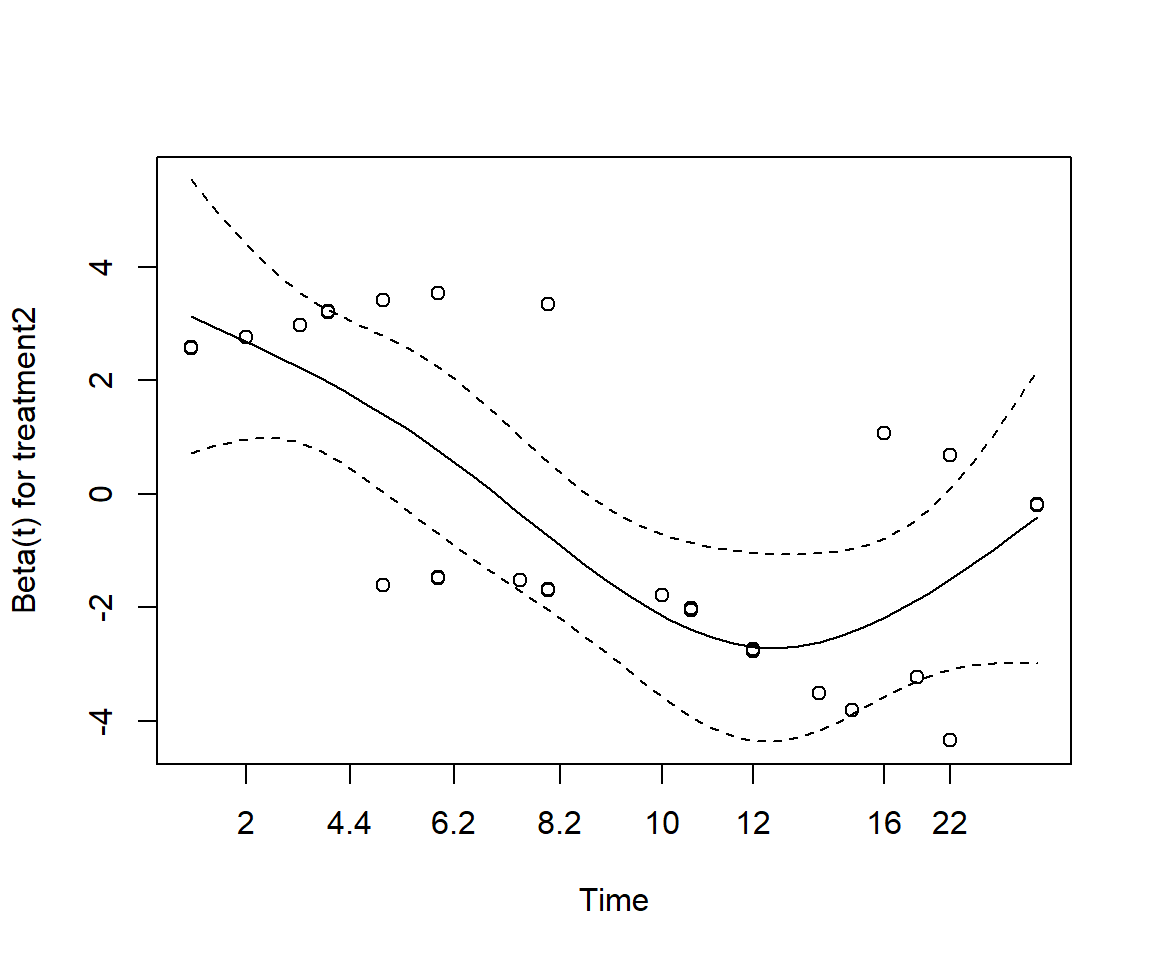
Perform a Cox regression of treatment2 and wbc3cat. Does Drug B have a significant effect on survival?
cox_adj2 <- coxph(Surv(weeks, relapse) ~ treatment2 + wbc3cat,
data = leukaemia)
exp(confint(cox_adj2))
2.5 % 97.5 %
treatment2Drug B 0.3706959 1.865371
wbc3catModerate 1.0869784 11.528659
wbc3catHigh 4.4682805 53.855488The effect of Drug B on survival is not statistically significant (the hazard ratio’s 95% confidence interval contains 1).
Test the proportional hazards assumption using the Schoenfeld residuals.
cox.zph(cox_adj2)
chisq df p
treatment2 18.216 1 2e-05
wbc3cat 0.268 2 0.87478
GLOBAL 19.590 3 0.00021The proportional hazards assumption does not hold overall; specifically not for treatment 2.
If you prefer visual tests:
The Kaplan-Meier curves suggest that Drug B has a negative effect on survival initially, then becomes positive. So we will test for different effects before and after 10 weeks. Generate a life table.
summary(surv_t2)
Call: survfit(formula = Surv(weeks, relapse) ~ treatment2, data = leukaemia)
treatment2=Standard
time n.risk n.event survival std.err lower 95% CI upper 95% CI
5 22 1 0.955 0.0444 0.8714 1.000
6 21 2 0.864 0.0732 0.7315 1.000
7 18 1 0.816 0.0834 0.6676 0.997
8 17 3 0.672 0.1020 0.4988 0.905
10 13 1 0.620 0.1064 0.4429 0.868
11 11 2 0.507 0.1131 0.3278 0.785
12 8 2 0.380 0.1150 0.2104 0.688
13 6 1 0.317 0.1119 0.1587 0.633
15 5 1 0.254 0.1060 0.1118 0.575
17 4 1 0.190 0.0966 0.0703 0.515
22 1 1 0.000 NaN NA NA
treatment2=Drug B
time n.risk n.event survival std.err lower 95% CI upper 95% CI
1 20 2 0.900 0.0671 0.778 1.000
2 18 2 0.800 0.0894 0.643 0.996
3 16 1 0.750 0.0968 0.582 0.966
4 15 2 0.650 0.1067 0.471 0.897
5 13 1 0.600 0.1095 0.420 0.858
6 12 1 0.550 0.1112 0.370 0.818
8 11 1 0.500 0.1118 0.323 0.775
16 10 1 0.450 0.1112 0.277 0.731
22 8 1 0.394 0.1106 0.227 0.683
23 7 2 0.281 0.1038 0.136 0.580Now, for each subject followed for more than 10 weeks, we will split the data into 2 observations, one for the time up to 10 weeks and one for the time after.
There is a good tutorial here explaining the approach of using step functions for time-varying coefficients. We can use the function survSplit() for this.
Has the life table changed?
Call: survfit(formula = Surv(tstart, weeks, relapse) ~ treatment2,
data = leuk2)
treatment2=Standard
time n.risk n.event survival std.err lower 95% CI upper 95% CI
5 22 1 0.955 0.0444 0.8714 1.000
6 21 2 0.864 0.0732 0.7315 1.000
7 18 1 0.816 0.0834 0.6676 0.997
8 17 3 0.672 0.1020 0.4988 0.905
10 13 1 0.620 0.1064 0.4429 0.868
11 11 2 0.507 0.1131 0.3278 0.785
12 8 2 0.380 0.1150 0.2104 0.688
13 6 1 0.317 0.1119 0.1587 0.633
15 5 1 0.254 0.1060 0.1118 0.575
17 4 1 0.190 0.0966 0.0703 0.515
22 1 1 0.000 NaN NA NA
treatment2=Drug B
time n.risk n.event survival std.err lower 95% CI upper 95% CI
1 20 2 0.900 0.0671 0.778 1.000
2 18 2 0.800 0.0894 0.643 0.996
3 16 1 0.750 0.0968 0.582 0.966
4 15 2 0.650 0.1067 0.471 0.897
5 13 1 0.600 0.1095 0.420 0.858
6 12 1 0.550 0.1112 0.370 0.818
8 11 1 0.500 0.1118 0.323 0.775
16 10 1 0.450 0.1112 0.277 0.731
22 8 1 0.394 0.1106 0.227 0.683
23 7 2 0.281 0.1038 0.136 0.580No.
Examine the data. You should see that for subjects who were followed up for less than ten weeks, there is a single record. However, for those followed up for more than 10 weeks, there are two records: one with tstart == 0 and one with tstart == 10.
Look at leuk2 (or whatever you decided to call the output of survSplit).
Now we can generate separate treatment variables for the treatment effect before and after 10 weeks.
This has already been done for us with the episode argument of survSplit. We have a variable called time_group.
Now fit the Cox regression model with these predictors. What is the hazard ratio for [drug B before 10 weeks] with its 95% confidence interval?
This involves an interaction between the time_group strata and treatment2. (Explained here.)
cox_split <- coxph(Surv(tstart, weeks, relapse) ~ wbc3cat + treatment2 * strata(time_group), data = leuk2)
exp(cbind(HR = coef(cox_split), confint(cox_split)))
HR
wbc3catModerate 3.14924983
wbc3catHigh 16.30334533
treatment2Drug B 2.18291244
treatment2Drug B:strata(time_group)time_group=2 0.04539226
2.5 %
wbc3catModerate 0.953390004
wbc3catHigh 4.631285085
treatment2Drug B 0.851712036
treatment2Drug B:strata(time_group)time_group=2 0.006887743
97.5 %
wbc3catModerate 10.4026416
wbc3catHigh 57.3920768
treatment2Drug B 5.5947392
treatment2Drug B:strata(time_group)time_group=2 0.2991483What is the hazard ratio for t2?
See above.
Do these hazard ratios confirm what you were expecting?
Yes, that Drug B increased risk initially, then reduced it.
Now test the proportional hazards assumption, using Schoenfeld residuals. Is the model now appropriate?
cox.zph(cox_split) %T>% plot
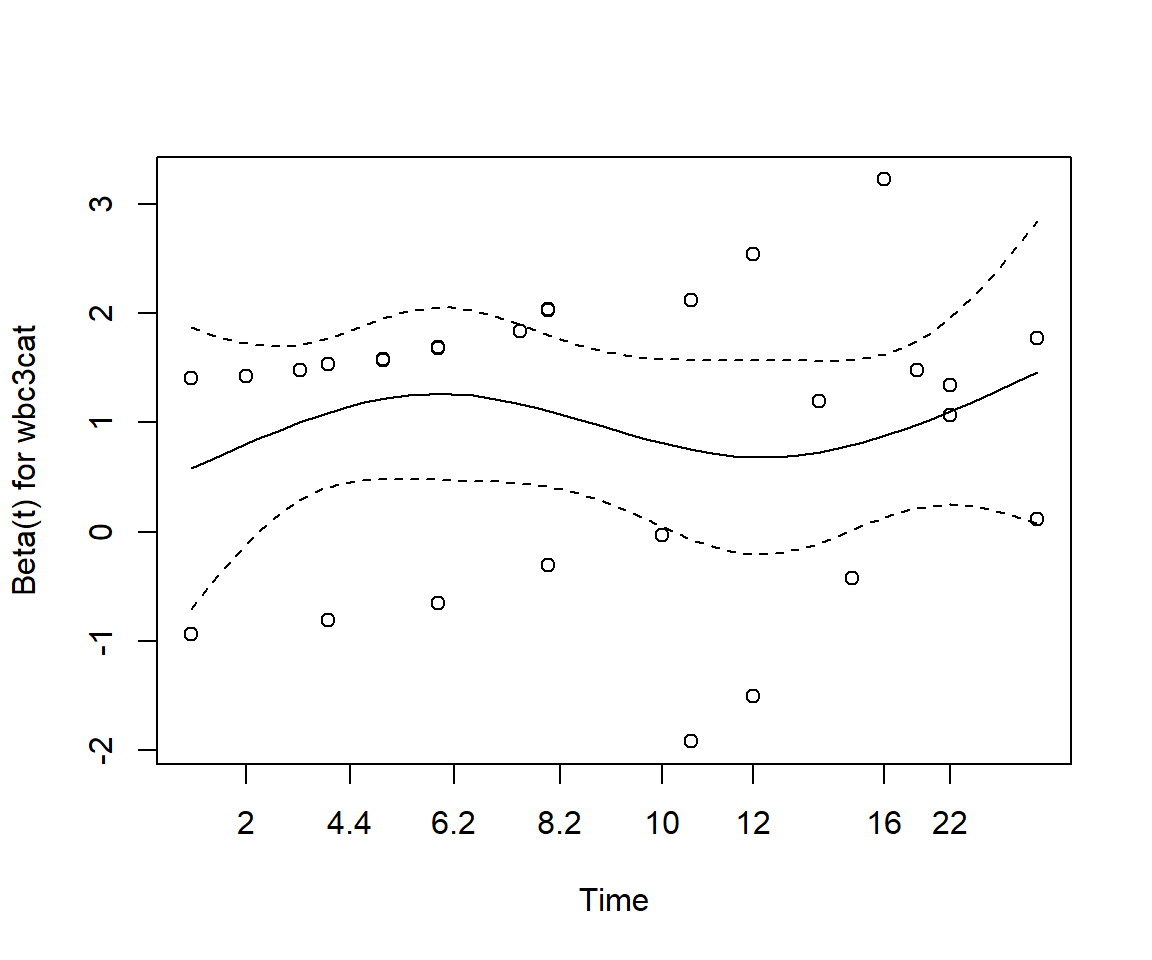
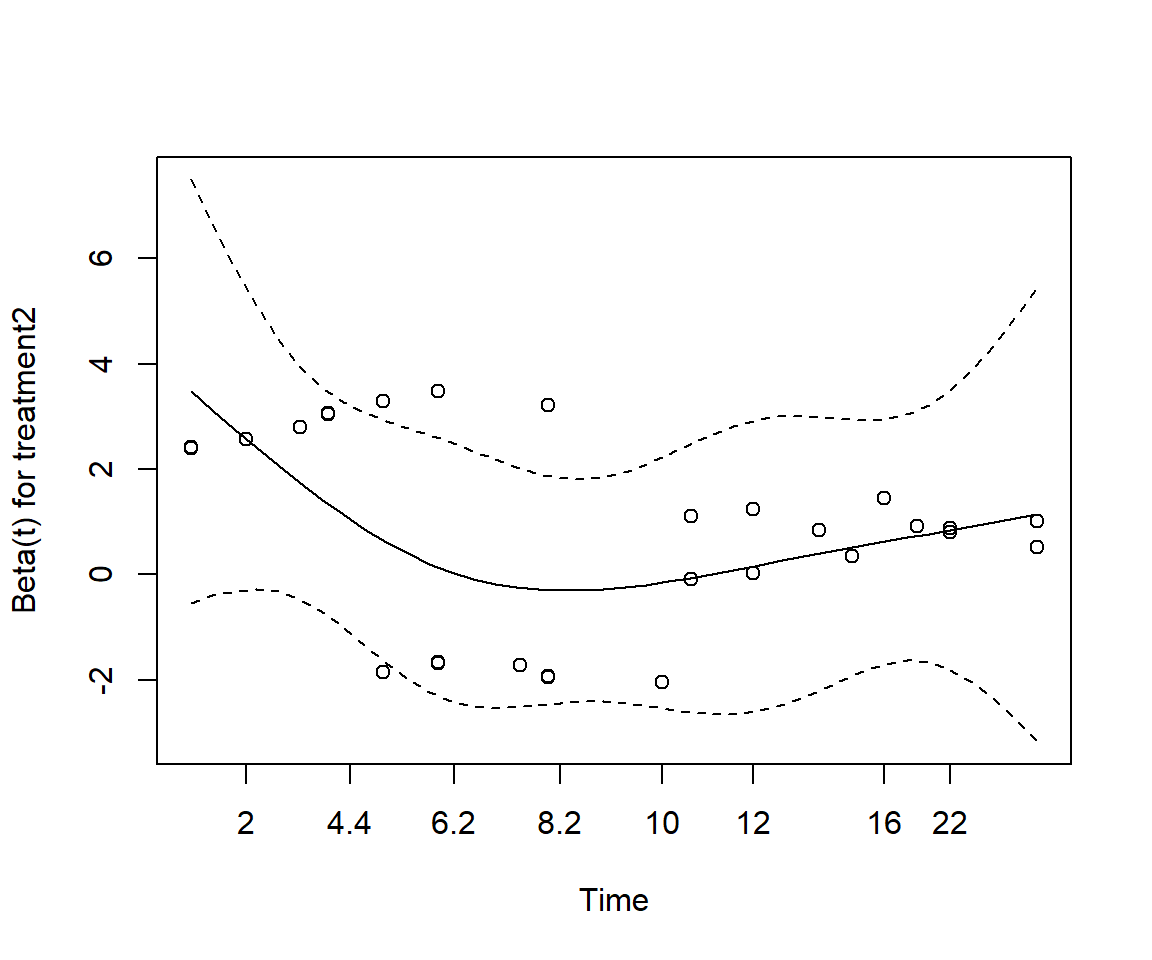
chisq df p
wbc3cat 1.540 2 0.463
treatment2 4.637 1 0.031
treatment2:strata(time_group) 0.736 1 0.391
GLOBAL 9.100 4 0.059The model is now appropriate and none of the predictors appear to show non-proportionality.