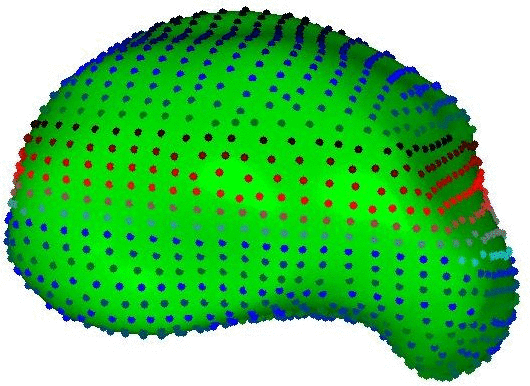
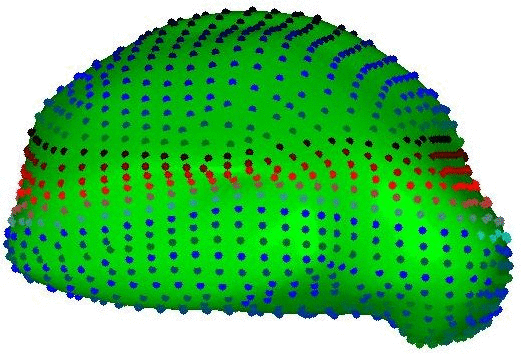
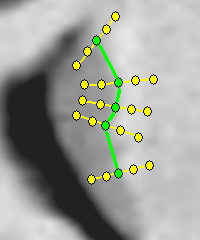
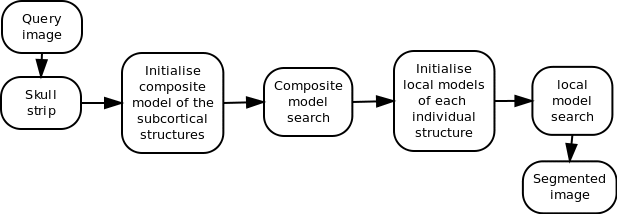
Integrated Brain Image modelling (IBIM)
|
Research Associate: Kola Babalola Principal Investigator: Tim Cootes Project Period: September 2004 - October 2007 Contents |
|
|
| ||||||||||||||||||||||
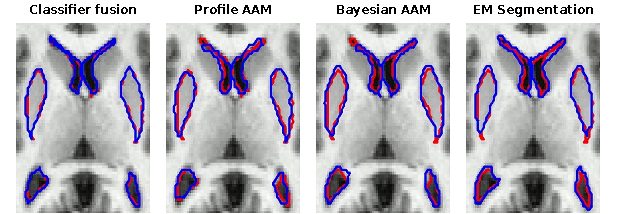
Selected publicationsAn evaluation of four automatic methods of segmenting the subcortical structures in the brain. KO Babalola, B Patenaude, P Aljabar, J Schnabel, D Kennedy, W Crum, S Smith, T Cootes, M Jenkinson, D Rueckert. Neuroimage, vol. 47, pp. 1435-1447, October 2009, doi:10.1016/j.neuroimage.2009.05.029 Comparison and Evaluation of Segmentation Techniques for Subcortical Structures in Brain MRI. KO Babalola, B Patenaude, P Aljabar, J Schnabel, D Kennedy, W Crum, S Smith, TF Cootes, M Jenkinson, and D Rueckert. In Proceedings of the 11th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), Lecture Notes in Computer Science, vol. 5241, pp. 409-416, September 2008, doi: 10.1007/978-3-540-85988-8_49 Comparing the Similarity of Statistical Shape Models using the Bhattacharya Metric. KO Babalola, TF Cootes, B Patenaude, A Rao, and M Jenkinson. In Proceedings of the 9th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), Lecture Notes in Computer Science, vol. 4190, pp. 142-150, October 2006, doi: 10.1007/11866565_18 Cannonical Correlation Analysis of Sub-cortical Brain Structures Using Non-rigid Registration. A Rao, Kolawole Babalola, and Daniel Rueckert. In Proceedings of the 3rd International Workshop on Biomedical Image Registration (WBIR), Lecture Notes in Computer Science, vol. 4057, pp. 66-74, July 2006, doi: 10.1007/11784012_9 Registering Richly Labelled 3D Images. KO Babalola and TF Cootes. In Proceedings of the International Symposium on Biomedical Images, Washington, Virginia, USA, April 2006, 10.1109/ISBI.2006.1625056 Back to contents |