Yassar Almutairi and Tim Cootes
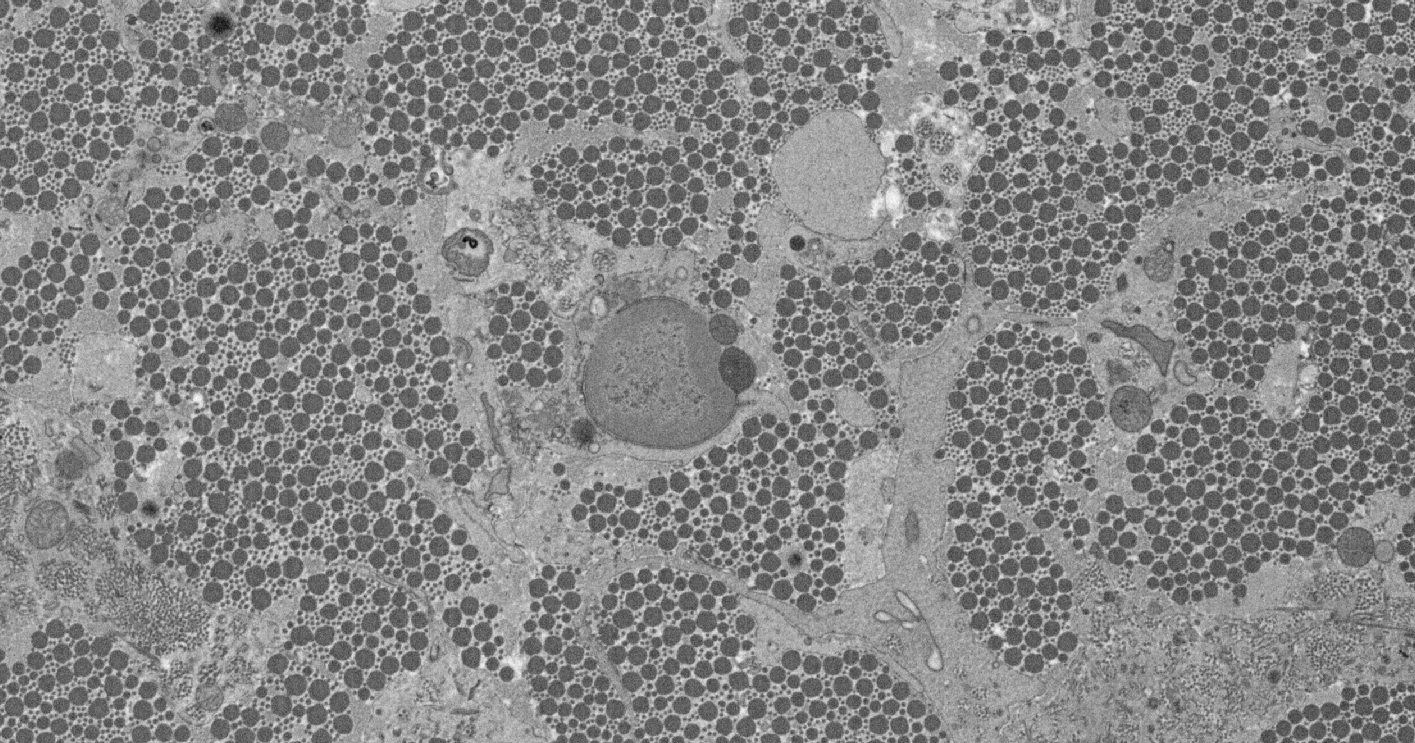
The goal of this project is to develop automatic or semi-automatic techniques to identify and classify a variety of structures in 3d images gathered using scanning electron microscopes. Such tools would allow biologists to examine the shape of cells and the matrix surrounding the cells during tissue development. One particular focus will be examining the tendon, which contains long collagen fibrils arranged in parallel bundles. Measuring the nature of the tendon structure would help examine how cells, that are microns in length, can organise a matrix of collagen fibrils that are only a few nanometres in diameter, but which can be millimetres long.
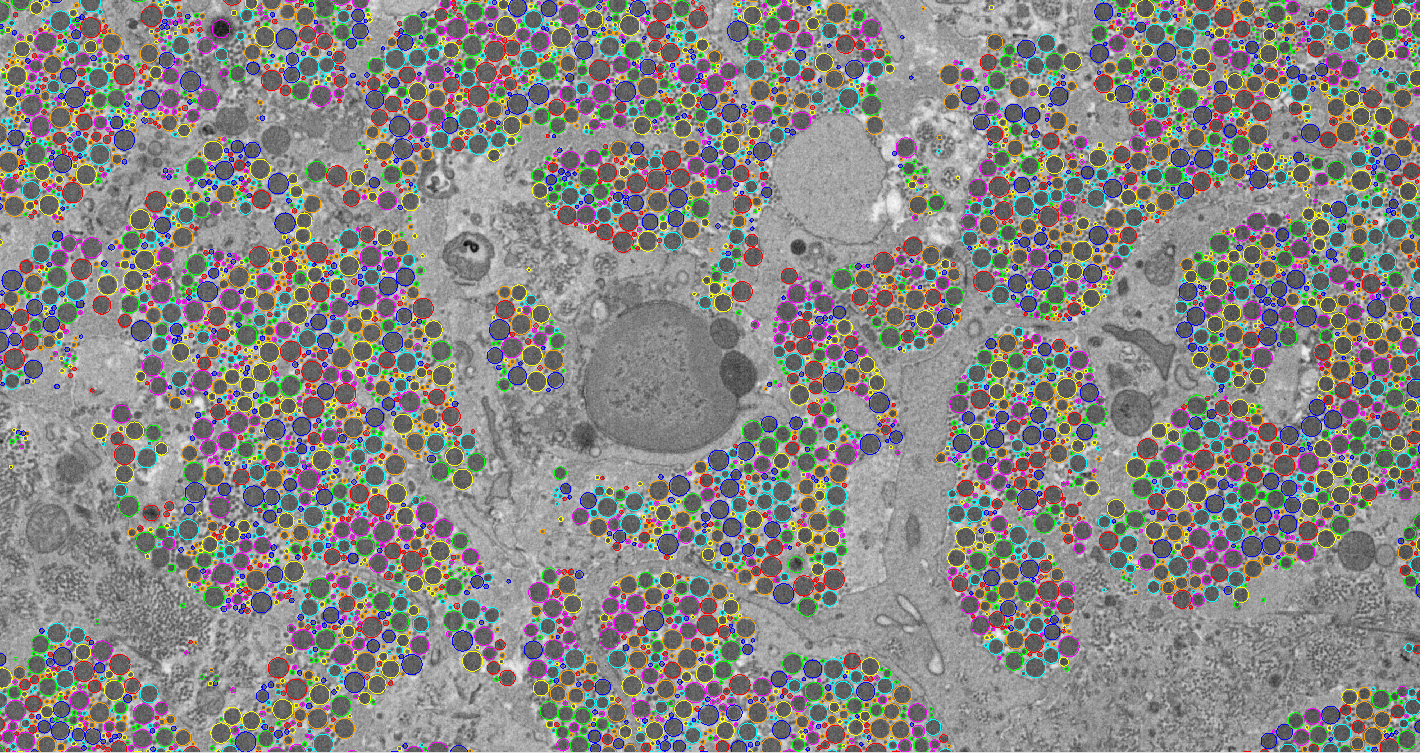
Currently we have identified and tracked fibres through the image volume. There can be over 30,000 fibres in a single image, and ideally each would be tracked through the volume in order to estimate their lengths and how they form into bundles. Manual annotation of such data has proved impractical. Our goal of this project is to create an automated system which detects the fibres and tracks them through large image volumes. We show that multi-scale normalised cross correlation is effective for finding candidates in a single image, and that false positive matches can be eliminated using a random forest classifier. Tracking is performed by linking candidates from one image to the next.
 |
 |
 |
 |
Thanks to Karl Kadler and Tobias Starborg for providing the images.
Y. Almutairi, T. F. Cootes, and K. Kadler. "Tracking collagen fibres through image volumes from SBFSEM". In Medical Image Understanding and Analysis - MIUA 2015. 19th Annual Conference, University of Lincoln, Lincoln, UK, July 15-17, 2015., pages 40-45, 2015. 2 (PDF)