Cluster validation
Cluster validation is concerned with the quality of clusters generated by an algorithm for data clustering. Given the partitioning of a data set, it attempts to answer questions such as: How pronounced is the cluster structure that has been identified? How do clustering solutions from different algorithms compare? How do clustering solutions for different parameters (e.g. the number of clusters compare).When information about true class memberships is available, external cluster validation techniques can be used, which provide an objective way of assessing algorithm performance. When such external knowledge is not available, internal measures need to be used, which attempt to measure the quality of the clusters based on the intrinsic properties of the data. Both approaches can be problematic, if biases of these methods are not taken into account. These issues are explored in detail in our 2006 Bioinformatics paper.
Our multiobjective clustering algorithm MOCK combines clustering and cluster validation.
Publications:
- Julia Handl, Joshua Knowles and Douglas Kell. (2005) Computational cluster validation in post-genomic data analysis. Bioinformatics 21(15):3201-3212
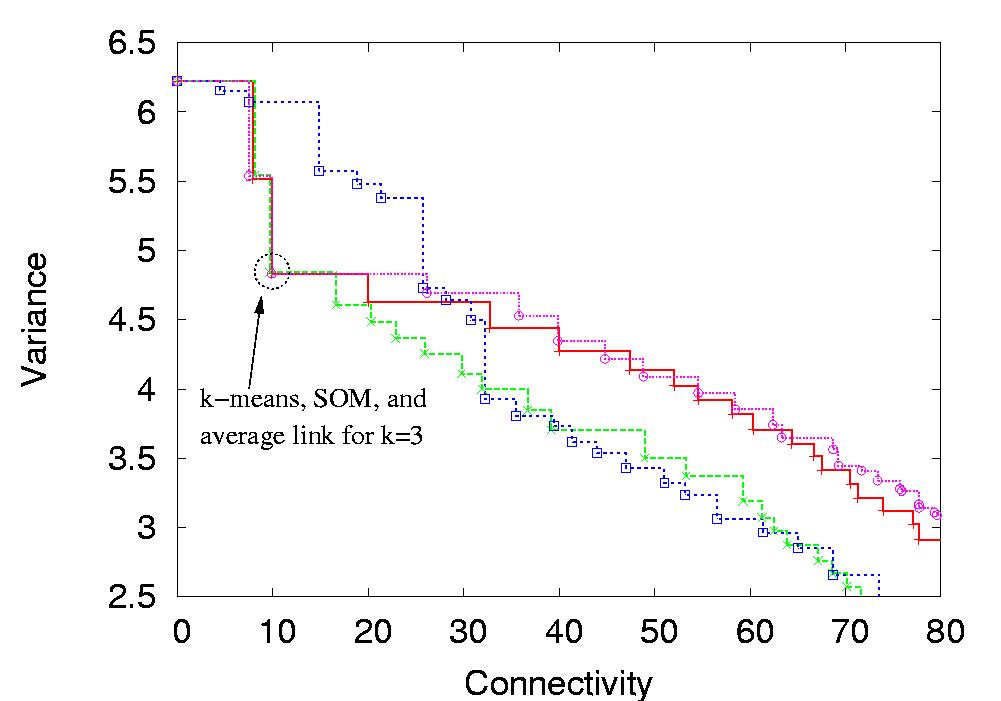
Figure 1: Visualization of the solutions from k-means, SOM, average link and single link in two-objective space. The knee corresponding to the "optimal" three-cluster solution on this data set is clearly pronounced. The visualization also shows the consistency between the k-means, SOM and average link solutions for k=2 and k=3, which further increases the confidence in the correctness of these partitionings.