Resources
- View the talk slides (source code)
- Stan web site
- JAGS (Just Another Gibbs Sampler)
- Statistical Rethinking book by Richard McElreath
- Sleep deprivation study
- original paper
- access the dataset in R with
data(sleepstudy, package = 'lme4')
- brms: Bayesian regression modelling with Stan
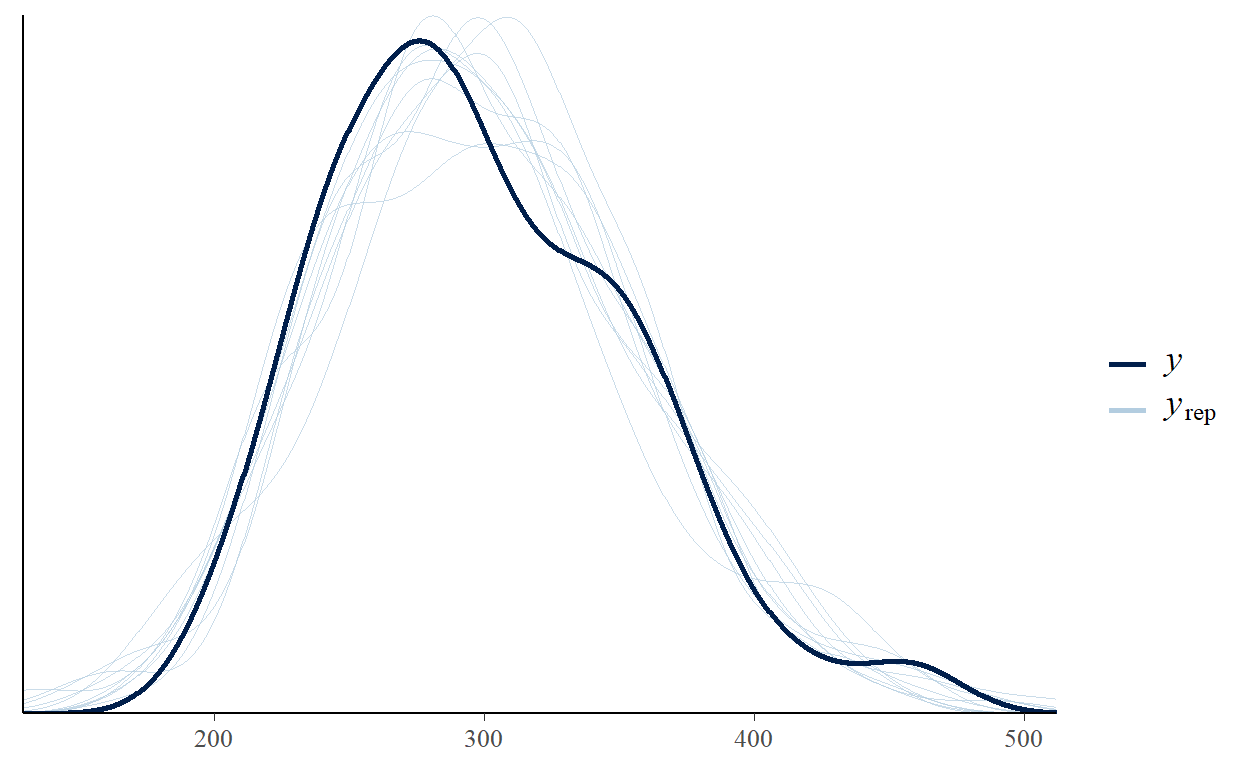
Final linear mixed model
fit <- brm(Reaction ~ Days + (Days | Subject),
data = lme4::sleepstudy)
Family: gaussian
Links: mu = identity; sigma = identity
Formula: Reaction ~ Days + (Days | Subject)
Data: lme4::sleepstudy (Number of observations: 180)
Samples: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup samples = 4000
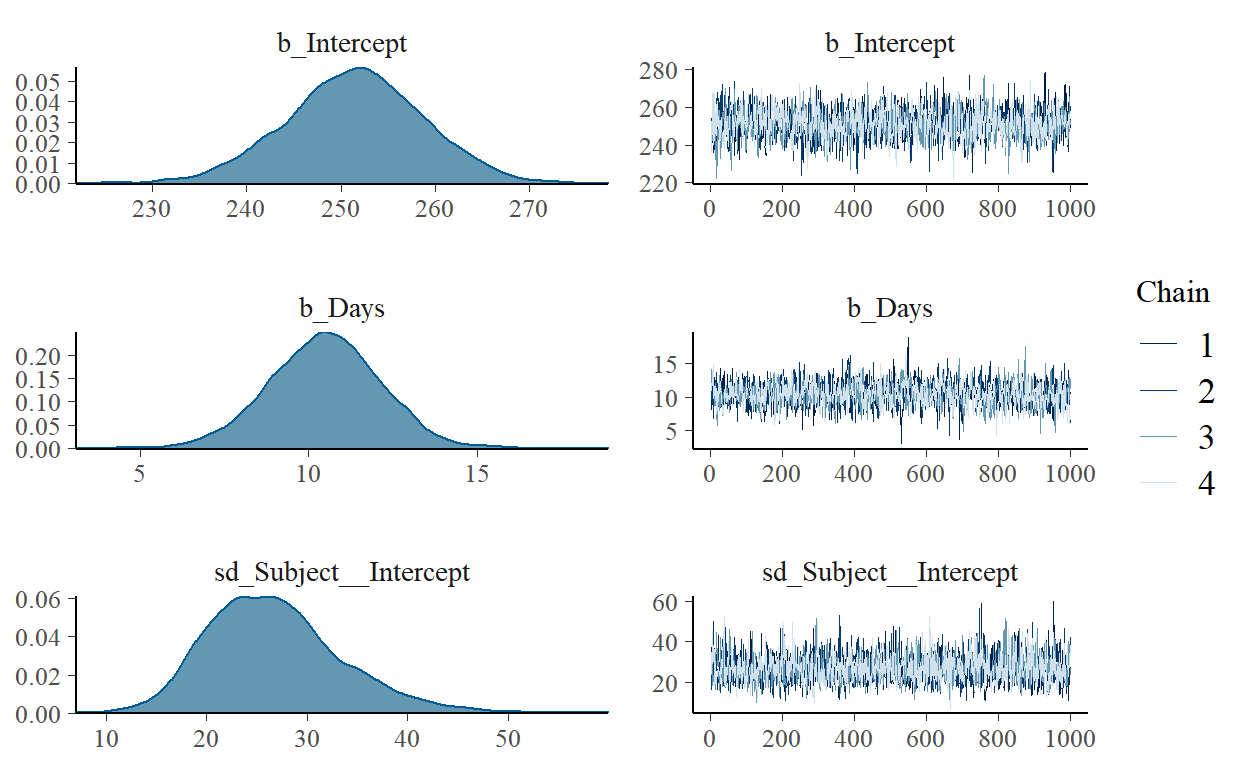
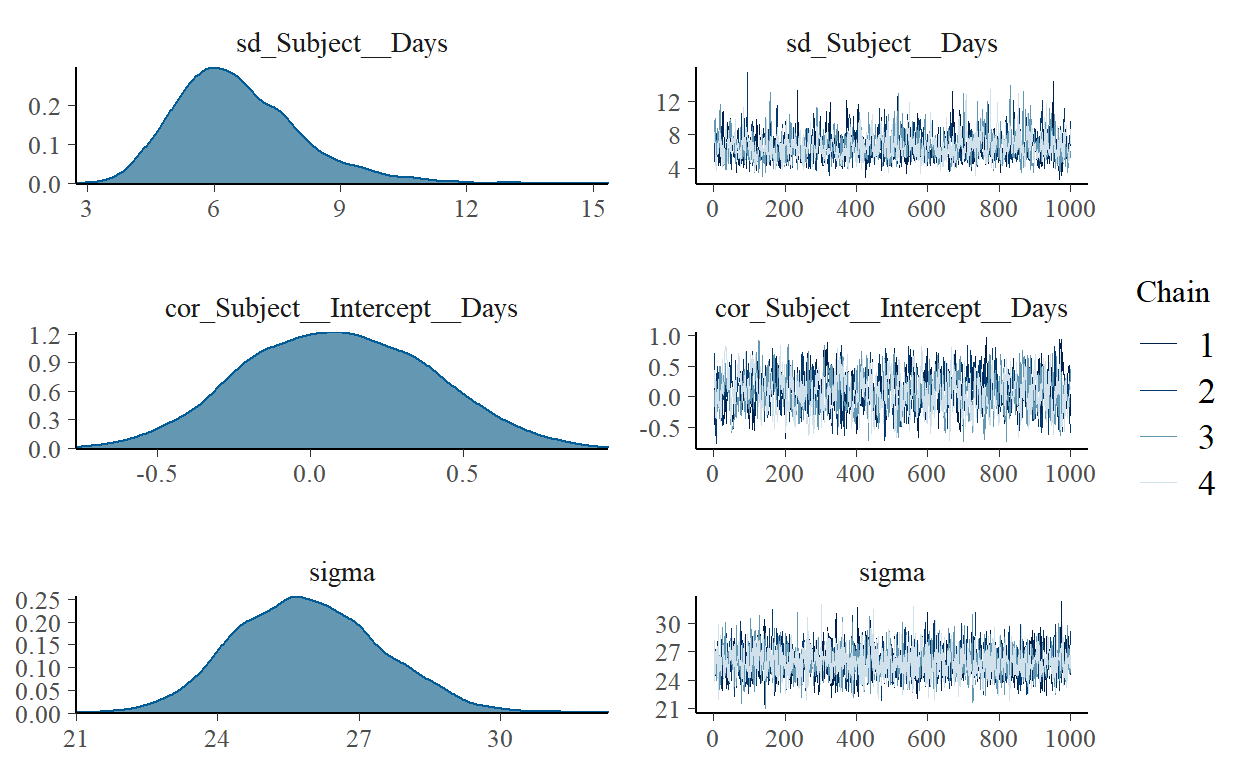
Group-Level Effects:
~Subject (Number of levels: 18)
Estimate Est.Error l-95% CI u-95% CI Rhat
sd(Intercept) 26.76 6.69 15.68 41.77 1.00
sd(Days) 6.58 1.51 4.22 10.11 1.00
cor(Intercept,Days) 0.09 0.30 -0.48 0.68 1.00
Bulk_ESS Tail_ESS
sd(Intercept) 2034 2769
sd(Days) 1635 1840
cor(Intercept,Days) 877 1633
Population-Level Effects:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
Intercept 251.45 7.45 236.65 265.67 1.00 1559 2236
Days 10.48 1.67 7.20 13.68 1.00 1385 2114
Family Specific Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma 25.92 1.55 23.10 29.02 1.00 3193 2917
Samples were drawn using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).