Motivating examples
By James Gwinnutt and Stephanie Shoop-Worrall
Computing summary statistics with tidyverse
The iris dataset is built into R and gives the measured widths and lengths of sepals and petals for 150 examples of three different species of iris.
Let’s compute some ‘baseline’ summary statistics for these data using the package dplyr (part of the tidyverse family of packages).
For a single variable, such as the Petal.Length, we can calculate species-level statistics as follows.
data(iris)
library(tidyverse)
iris %>%
group_by(Species) %>%
summarise(mean = mean(Petal.Length),
sd = sd(Petal.Length),
n = n(),
median = median(Petal.Length),
Q1 = quantile(Petal.Length, .25),
Q3 = quantile(Petal.Length, .75),
IQR = IQR(Petal.Length),
IQR1 = Q3 - Q1) # just another way of calculating iqr
# A tibble: 3 x 9
Species mean sd n median Q1 Q3 IQR IQR1
<fct> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
1 setosa 1.46 0.174 50 1.5 1.4 1.58 0.175 0.175
2 versicolor 4.26 0.470 50 4.35 4 4.6 0.600 0.600
3 virginica 5.55 0.552 50 5.55 5.1 5.88 0.775 0.775If we want to compute the same summaries for the other variables: petal width, sepal width and sepal length, then we could just repeat the code above, swapping the name Petal.Length each time. But there is a better, less repetitive way.
Using another tidyverse package called tidyr, we can reshape the dataset into a long (rather than wide) table.
(iris_long <- iris %>%
pivot_longer(-Species))
# A tibble: 600 x 3
Species name value
<fct> <chr> <dbl>
1 setosa Sepal.Length 5.1
2 setosa Sepal.Width 3.5
3 setosa Petal.Length 1.4
4 setosa Petal.Width 0.2
5 setosa Sepal.Length 4.9
6 setosa Sepal.Width 3
7 setosa Petal.Length 1.4
8 setosa Petal.Width 0.2
9 setosa Sepal.Length 4.7
10 setosa Sepal.Width 3.2
# ... with 590 more rowsInstead of a separate column for each type of measurement, we have one column for the variable names and one for the values. Then the name can be just another group to pass to group_by:
iris_long %>%
group_by(Species, Variable = name) %>%
summarise(mean = mean(value),
sd = sd(value),
n = n(),
median = median(value),
Q1 = quantile(value, .25),
Q3 = quantile(value, .75),
IQR = IQR(value))
# A tibble: 12 x 9
# Groups: Species [3]
Species Variable mean sd n median Q1 Q3 IQR
<fct> <chr> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <dbl>
1 setosa Petal.Length 1.46 0.174 50 1.5 1.4 1.58 0.175
2 setosa Petal.Width 0.246 0.105 50 0.2 0.2 0.3 0.100
3 setosa Sepal.Length 5.01 0.352 50 5 4.8 5.2 0.4
4 setosa Sepal.Width 3.43 0.379 50 3.4 3.2 3.68 0.475
5 versicolor Petal.Length 4.26 0.470 50 4.35 4 4.6 0.600
6 versicolor Petal.Width 1.33 0.198 50 1.3 1.2 1.5 0.3
7 versicolor Sepal.Length 5.94 0.516 50 5.9 5.6 6.3 0.7
8 versicolor Sepal.Width 2.77 0.314 50 2.8 2.52 3 0.475
9 virginica Petal.Length 5.55 0.552 50 5.55 5.1 5.88 0.775
10 virginica Petal.Width 2.03 0.275 50 2 1.8 2.3 0.500
11 virginica Sepal.Length 6.59 0.636 50 6.5 6.22 6.9 0.675
12 virginica Sepal.Width 2.97 0.322 50 3 2.8 3.18 0.375From here, see the last section on how we might turn this into a publication-quality table like below.
| Mean | SD | Median | Q1 | Q3 | IQR | |
|---|---|---|---|---|---|---|
| setosa (N = 50) | ||||||
| Petal Length | 1.5 | 0.2 | 1.5 | 1.4 | 1.6 | 0.2 |
| Petal Width | 0.2 | 0.1 | 0.2 | 0.2 | 0.3 | 0.1 |
| Sepal Length | 5.0 | 0.4 | 5.0 | 4.8 | 5.2 | 0.4 |
| Sepal Width | 3.4 | 0.4 | 3.4 | 3.2 | 3.7 | 0.5 |
| versicolor (N = 50) | ||||||
| Petal Length | 4.3 | 0.5 | 4.3 | 4.0 | 4.6 | 0.6 |
| Petal Width | 1.3 | 0.2 | 1.3 | 1.2 | 1.5 | 0.3 |
| Sepal Length | 5.9 | 0.5 | 5.9 | 5.6 | 6.3 | 0.7 |
| Sepal Width | 2.8 | 0.3 | 2.8 | 2.5 | 3.0 | 0.5 |
| virginica (N = 50) | ||||||
| Petal Length | 5.6 | 0.6 | 5.6 | 5.1 | 5.9 | 0.8 |
| Petal Width | 2.0 | 0.3 | 2.0 | 1.8 | 2.3 | 0.5 |
| Sepal Length | 6.6 | 0.6 | 6.5 | 6.2 | 6.9 | 0.7 |
| Sepal Width | 3.0 | 0.3 | 3.0 | 2.8 | 3.2 | 0.4 |
Recoding variables
Sometimes variables aren’t quite in the data type that we want. For example, we might want to split a continuous variable like BMI into categories based on pre-specified cutpoints. Suppose our vector of BMIs was
bmi <- c(29.4, 11.4, 24.5, 19.1, -0.6, 7.9, NA, 20.1, 30, 40.9)
And the pre-specified groups boundaries are at 0, 18.5, 25, 30, 35 and 40. One way of doing this would be a series of nested ifelse() expressions.
ifelse(bmi >= 0 & bmi < 18.5, '[0, 18.5)',
ifelse(bmi >= 18.5 & bmi < 25, '[18.5, 25)',
ifelse(bmi >= 25 & bmi < 30, '[25, 30)',
ifelse(bmi >= 30 & bmi < 35, '[30, 35)',
ifelse(bmi >= 35 & bmi < 40, '[35, 40)',
ifelse(bmi >= 40, '40+', NA))))))
[1] "[25, 30)" "[0, 18.5)" "[18.5, 25)" "[18.5, 25)" NA
[6] "[0, 18.5)" NA "[18.5, 25)" "[30, 35)" "40+" However, this is tedious to write, error-prone and unnecessarily verbose. There are built-in functions to do this! One is called cut() and the other is called findInterval(). The former creates a factor variable, labelled according to the interval in which each value falls. The latter is similar but just returns an integer corresponding to the interval.
[1] (25,30] (0,18.5] (18.5,25] (18.5,25] <NA> (0,18.5]
[7] <NA> (18.5,25] (25,30] <NA>
Levels: (0,18.5] (18.5,25] (25,30] (30,35] (35,40]By default, anything greater than the last cutpoint (or smaller than the first), if passed to cut(), is categorised as NA. To avoid this happening, you can set the first and last cutpoints to arbitrarily small and high numbers, respectively (such as -Inf and +Inf).
Negative BMIs are clearly invalid but let’s try to keep the 40+ ones. To match the first example, let’s also have the intervals open (exclusive) on the right:
[1] [25,30) [0,18.5) [18.5,25) [18.5,25) <NA> [0,18.5)
[7] <NA> [18.5,25) [30,35) [40,Inf)
Levels: [0,18.5) [18.5,25) [25,30) [30,35) [35,40) [40,Inf)Alternatively, the related function findInterval() will classify anything before the first cutpoint as group 0 and anything after the last cutpoint as group \(n_\text{cutpoints}+1\).
findInterval(bmi, cutpoints)
[1] 3 1 2 2 0 1 NA 2 4 6Explore the documentation for both functions to see what other options are available.
Now let’s use what we’ve learnt to count the number of patients in each group. Suppose the data are stored in a data frame called bmi_data:
id bmi
1 1 29.4
2 2 11.4
3 3 24.5
4 4 19.1
5 5 -0.6
6 6 7.9
7 7 NA
8 8 20.1
9 9 30.0
10 10 40.9Then a dplyr approach might be
bmi_data %>%
mutate(bmi_group = findInterval(bmi, cutpoints)) %>%
count(bmi_group)
bmi_group n
1 0 1
2 1 2
3 2 3
4 3 1
5 4 1
6 6 1
7 NA 1Here’s a base R solution, using automatic labels and treating negative BMIs as invalid.
(0,18.5] (18.5,25] (25,30] (30,35] (35,40] (40,Inf]
2 3 2 0 0 1 table(bmi_data$bmi_group, useNA = 'ifany')
(0,18.5] (18.5,25] (25,30] (30,35] (35,40] (40,Inf] <NA>
2 3 2 0 0 1 2 Merging categories
Imagine we want to merge categories that have fewer than 10 people in them, for confidentiality purposes. There are many ways you might do this.
example_data
# A tibble: 500 x 3
id country score
<int> <chr> <dbl>
1 1 Spain 14
2 2 Ireland 17
3 3 Montenegro 29
4 4 Portugal 21
5 5 Romania 25
6 6 Latvia 13
7 7 Isle of Man 23
8 8 Guernsey 10
9 9 Slovenia 21
10 10 Andorra 25
# ... with 490 more rowsOne way is to add_counts for each group, then mutate using ifelse. Here I am creating a new column called new_country so you can compare them.
example_data %>%
add_count(country, name = 'N') %>%
mutate(new_country = ifelse(N < 10, 'Other', country))
# A tibble: 500 x 5
id country score N new_country
<int> <chr> <dbl> <int> <chr>
1 1 Spain 14 16 Spain
2 2 Ireland 17 13 Ireland
3 3 Montenegro 29 7 Other
4 4 Portugal 21 7 Other
5 5 Romania 25 11 Romania
6 6 Latvia 13 9 Other
7 7 Isle of Man 23 11 Isle of Man
8 8 Guernsey 10 8 Other
9 9 Slovenia 21 11 Slovenia
10 10 Andorra 25 13 Andorra
# ... with 490 more rowsAlternatively, you might use the fct_lump_min (factor lump) function from the forcats (“for categorical variables”) package, which is specifically designed for this sort of task. There are lots of other fun functions for manipulating categorical variables in that package; check it out!
example_data %>%
mutate(new_country = forcats::fct_lump_min(country, min = 10))
# A tibble: 500 x 4
id country score new_country
<int> <chr> <dbl> <fct>
1 1 Spain 14 Spain
2 2 Ireland 17 Ireland
3 3 Montenegro 29 Other
4 4 Portugal 21 Other
5 5 Romania 25 Romania
6 6 Latvia 13 Other
7 7 Isle of Man 23 Isle of Man
8 8 Guernsey 10 Other
9 9 Slovenia 21 Slovenia
10 10 Andorra 25 Andorra
# ... with 490 more rowsOr if you prefer data.table, this solution is analogous to using dplyr::add_count. The row-indexing syntax saves us a call to ifelse (or fifelse or if_else…).
library(data.table)
setDT(example_data)
example_data[, N := .N, by = country]
example_data[N < 10, country := 'Other']
example_data
id country score N
1: 1 Spain 14 16
2: 2 Ireland 17 13
3: 3 Other 29 7
4: 4 Other 21 7
5: 5 Romania 25 11
---
496: 496 Other 26 8
497: 497 Other 17 7
498: 498 Other 33 7
499: 499 Other 23 8
500: 500 Albania 16 11Counting missing values
How much missing data is there for each variable in the baseline table? Here’s an example using the airquality dataset (which is built into base R and contains some missing values).
Ozone Solar.R Wind Temp Month Day
1 41 190 7.4 67 5 1
2 36 118 8.0 72 5 2
3 12 149 12.6 74 5 3
4 18 313 11.5 62 5 4
5 NA NA 14.3 56 5 5
6 28 NA 14.9 66 5 6The base summary function will report NA counts, if any.
summary(airquality)
Ozone Solar.R Wind Temp
Min. : 1.00 Min. : 7.0 Min. : 1.700 Min. :56.00
1st Qu.: 18.00 1st Qu.:115.8 1st Qu.: 7.400 1st Qu.:72.00
Median : 31.50 Median :205.0 Median : 9.700 Median :79.00
Mean : 42.13 Mean :185.9 Mean : 9.958 Mean :77.88
3rd Qu.: 63.25 3rd Qu.:258.8 3rd Qu.:11.500 3rd Qu.:85.00
Max. :168.00 Max. :334.0 Max. :20.700 Max. :97.00
NA's :37 NA's :7
Month Day
Min. :5.000 Min. : 1.0
1st Qu.:6.000 1st Qu.: 8.0
Median :7.000 Median :16.0
Mean :6.993 Mean :15.8
3rd Qu.:8.000 3rd Qu.:23.0
Max. :9.000 Max. :31.0
Or you can loop over the columns and compute the counts and proportions:
airquality_nas <- airquality[, 1:4]
airquality_nas[] <- lapply(airquality_nas, is.na)
colSums(airquality_nas)
Ozone Solar.R Wind Temp
37 7 0 0 100 * colMeans(airquality_nas)
Ozone Solar.R Wind Temp
24.183007 4.575163 0.000000 0.000000 In dplyr, it might look something like this:
airquality %>%
pivot_longer(Ozone:Temp) %>% # ignore Day & Month
group_by(name) %>%
summarise(total = n(),
present = sum(!is.na(value)),
missing = sum(is.na(value)),
`% present` = 100 * present / total, # or mean(!is.na(value))
`% missing` = 100 * missing / total) # or mean(is.na(value))
# A tibble: 4 x 6
name total present missing `% present` `% missing`
<chr> <int> <int> <int> <dbl> <dbl>
1 Ozone 153 116 37 75.8 24.2
2 Solar.R 153 146 7 95.4 4.58
3 Temp 153 153 0 100 0
4 Wind 153 153 0 100 0 And in data.table:
setDT(airquality)
airquality[, lapply(.SD, is.na)][,
.(column = names(.SD),
total = .N,
missing = sapply(.SD, sum),
`% missing` = sapply(.SD, mean),
present = .N - sapply(.SD, sum),
`% present` = 1 - sapply(.SD, mean))]
column total missing % missing present % present
1: Ozone 153 37 0.24183007 116 0.7581699
2: Solar.R 153 7 0.04575163 146 0.9542484
3: Wind 153 0 0.00000000 153 1.0000000
4: Temp 153 0 0.00000000 153 1.0000000
5: Month 153 0 0.00000000 153 1.0000000
6: Day 153 0 0.00000000 153 1.0000000The TableOne package
By Ruth Costello
The R package tableone quickly produces a table that is easy to use in medical research papers. Given a specification of variables (vars), factors (factorVars) and groups (strata), it computes common summary statistics automatically.
For example:
library(tableone)
CreateTableOne(
vars = c('Petal.Length', 'Petal.Width'),
strata = 'Species',
data = iris
)
Stratified by Species
setosa versicolor virginica p test
n 50 50 50
Petal.Length (mean (SD)) 1.46 (0.17) 4.26 (0.47) 5.55 (0.55) <0.001
Petal.Width (mean (SD)) 0.25 (0.11) 1.33 (0.20) 2.03 (0.27) <0.001 Another example:
CreateTableOne(
vars = c('age', 'sex', 'state', 'T.categ'),
factorVars = c('state', 'sex', 'T.categ'),
data = MASS::Aids2
)
Overall
n 2843
age (mean (SD)) 37.41 (10.06)
sex = M (%) 2754 (96.9)
state (%)
NSW 1780 (62.6)
Other 249 ( 8.8)
QLD 226 ( 7.9)
VIC 588 (20.7)
T.categ (%)
hs 2465 (86.7)
hsid 72 ( 2.5)
id 48 ( 1.7)
het 41 ( 1.4)
haem 46 ( 1.6)
blood 94 ( 3.3)
mother 7 ( 0.2)
other 70 ( 2.5) Other, similar packages you might like try include:
- Package
TableOne(not on CRAN; based on this paper) - Package
table1 - Package
gtsummary
Neater knitr tables
By David Selby
Generating tables by hand
If copying data from an external source (rather than calculating it yourself), then you might simply want to write in the data by hand. For this you can use a Markdown table, for instance, the following Markdown pipe table is copied from the supplementary materials of Dixon et al. (2019) and annotated with a Markdown caption.
| Variable | n | % |
|---|---|---|
| Number of participants | 2658 | 100 |
| Female | 2210 | 83.1 |
| Rheumatoid arthritis | 506 | 19.0 |
| Osteoarthritis | 926 | 34.8 |
| Ankylosing spondylitis | 235 | 8.8 |
| Gout | 97 | 3.6 |
| Arthritis (unspecified) | 972 | 36.6 |
| Fibromyalgia | 665 | 25.0 |
| Chronic headache | 271 | 10.2 |
| Neuropathic pain | 371 | 14.0 |
using the following Markdown ‘pipe table’ syntax:
Table: Participants in the _Cloudy with a Chance of Pain_ study (Dixon et al, 2019)
| Variable | n | % |
| :---------------------- | ---: | ---: |
| Number of participants | 2658 | 100 |
| Female | 2210 | 83.1 |
| Rheumatoid arthritis | 506 | 19.0 |
| Osteoarthritis | 926 | 34.8 |
| Ankylosing spondylitis | 235 | 8.8 |
| Gout | 97 | 3.6 |
| Arthritis (unspecified) | 972 | 36.6 |
| Fibromyalgia | 665 | 25.0 |
| Chronic headache | 271 | 10.2 |
| Neuropathic pain | 371 | 14.0 |But you don’t need to space the syntax perfectly. This works just as well:
Variable | n | %
:--- | ---: | ---:
Number of participants | 2658 | 100
Female | 2210 | 83.1
Rheumatoid arthritis | 506 | 19.0
Osteoarthritis | 926 | 34.8
Ankylosing spondylitis | 235 | 8.8
Gout | 97 | 3.6
Arthritis (unspecified) | 972 | 36.6
Fibromyalgia | 665 | 25.0
Chronic headache | 271 | 10.2
Neuropathic pain | 371 | 14.0Which you can leave as-is, or you can neaten up automagically using the RStudio Addin Beautify Table from the beautifyR package. Install the addin from GitHub using the syntax
devtools::install_github('mwip/beautifyR')then highlight your Markdown table and click Addins > Beautify Table from the toolbar.
Suppose you want to do some calculations with the copied data—not just display it in a table. You could enter it into a data frame by hand, using base syntax or the (slightly easier to read) tibble::tribble function:
my_tbl <- tibble::tribble(
~Variable, ~n, ~`%`,
"Number of participants", 2658L, 100,
"Female", 2210L, 83.1,
"Rheumatoid arthritis", 506L, 19,
"Osteoarthritis", 926L, 34.8,
"Ankylosing spondylitis", 235L, 8.8,
"Gout", 97L, 3.6,
"Arthritis (unspecified)", 972L, 36.6,
"Fibromyalgia", 665L, 25,
"Chronic headache", 271L, 10.2,
"Neuropathic pain", 371L, 14
)
Thence, wrap the data frame in a call to kable, xtable or pander within a script or code chunk (or print it in an R Markdown code chunk with your YAML header set to df_print: kable).
knitr::kable(my_tbl)
pander::pander(my_tbl)
xtable::xtable(my_tbl)| Variable | n | % |
|---|---|---|
| Number of participants | 2658 | 100.0 |
| Female | 2210 | 83.1 |
| Rheumatoid arthritis | 506 | 19.0 |
| Osteoarthritis | 926 | 34.8 |
| Ankylosing spondylitis | 235 | 8.8 |
| Gout | 97 | 3.6 |
| Arthritis (unspecified) | 972 | 36.6 |
| Fibromyalgia | 665 | 25.0 |
| Chronic headache | 271 | 10.2 |
| Neuropathic pain | 371 | 14.0 |
Copy and paste
But manually typing in these values is still tedious and error-prone. Can’t we just copy and paste? Yes we can, with the datapasta package. Install the package:
install.packages('datapasta')Copy your source table to the clipboard (e.g. from a webpage or Word document) and then select Addins > Paste as (format) into code chunk or script. (I used it to generate the tribble code above!)
Complex kable tables
Functions like kable (Xie 2014) and pander are straightforward to use for simple tables, but do not have many advanced features. For that, try using the package kableExtra (Zhu 2020), which adds just about all the functionality you could ever really need.
Here are some examples.
library(kableExtra)
options(knitr.table.format = 'html')
Add header rows to group columns
kable(my_tbl) %>%
kable_styling() %>%
add_header_above(c(' ' = 1, 'Final case-crossover study cohort' = 2))
| Variable | n | % |
|---|---|---|
| Number of participants | 2658 | 100.0 |
| Female | 2210 | 83.1 |
| Rheumatoid arthritis | 506 | 19.0 |
| Osteoarthritis | 926 | 34.8 |
| Ankylosing spondylitis | 235 | 8.8 |
| Gout | 97 | 3.6 |
| Arthritis (unspecified) | 972 | 36.6 |
| Fibromyalgia | 665 | 25.0 |
| Chronic headache | 271 | 10.2 |
| Neuropathic pain | 371 | 14.0 |
Add indents to group rows implicitly
knitr::kable(my_tbl) %>%
add_indent(c(2, 4), level_of_indent = 1)
| Variable | n | % |
|---|---|---|
| Number of participants | 2658 | 100.0 |
| Female | 2210 | 83.1 |
| Rheumatoid arthritis | 506 | 19.0 |
| Osteoarthritis | 926 | 34.8 |
| Ankylosing spondylitis | 235 | 8.8 |
| Gout | 97 | 3.6 |
| Arthritis (unspecified) | 972 | 36.6 |
| Fibromyalgia | 665 | 25.0 |
| Chronic headache | 271 | 10.2 |
| Neuropathic pain | 371 | 14.0 |
Add labels to group rows explicitly
| Variable | n | % |
|---|---|---|
| Number of participants | 2658 | 100.0 |
| Demographics | ||
| Female | 2210 | 83.1 |
| Health conditions | ||
| Rheumatoid arthritis | 506 | 19.0 |
| Osteoarthritis | 926 | 34.8 |
| Ankylosing spondylitis | 235 | 8.8 |
| Gout | 97 | 3.6 |
| Arthritis (unspecified) | 972 | 36.6 |
| Fibromyalgia | 665 | 25.0 |
| Chronic headache | 271 | 10.2 |
| Neuropathic pain | 371 | 14.0 |
Collapse rows that are repeated
kable(engagement_days) %>%
kable_styling() %>%
collapse_rows(valign = 'top')
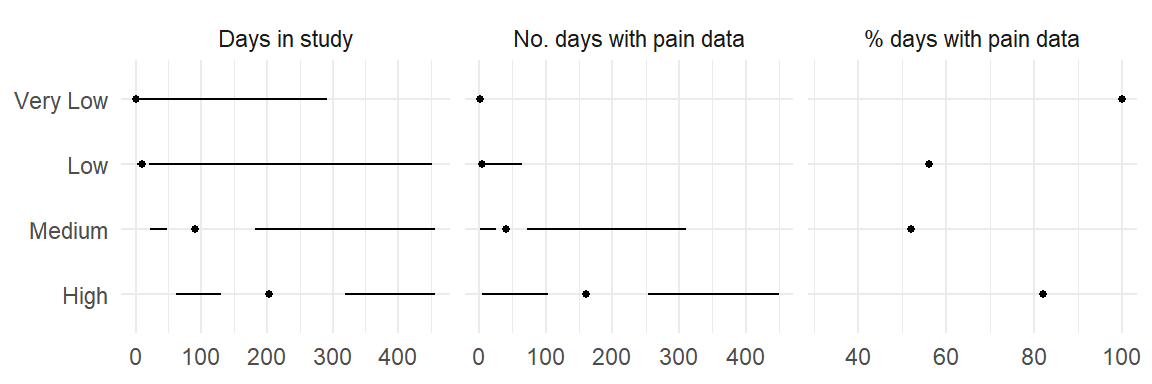
| Cluster | Engagement | Min | Q1 | Median | Q3 | Max |
|---|---|---|---|---|---|---|
| High | Days in study | 61 | 130 | 203 | 319 | 456 |
| No. days with pain data | 4 | 104 | 160 | 254 | 449 | |
| % days with pain data | 73 | 82 | 89 | |||
| Medium | Days in study | 22 | 48 | 91 | 182 | 456 |
| No. days with pain data | 2 | 26 | 40 | 72 | 311 | |
| % days with pain data | 38 | 52 | 67 | |||
| Low | Days in study | 2 | 4 | 10 | 21 | 452 |
| No. days with pain data | 1 | 2 | 5 | 9 | 64 | |
| % days with pain data | 32 | 56 | 81 | |||
| Very Low | Days in study | 1 | 1 | 1 | 1 | 291 |
| No. days with pain data | 2 | |||||
| % days with pain data | 100 | 100 | 100 |
library(dplyr)
## Rearrange and sort the columns:
engagement_days %>%
select(Engagement, Cluster, Min:Max) %>%
arrange(Engagement, Cluster) %>%
kable() %>%
kable_styling %>%
collapse_rows(valign = 'top')
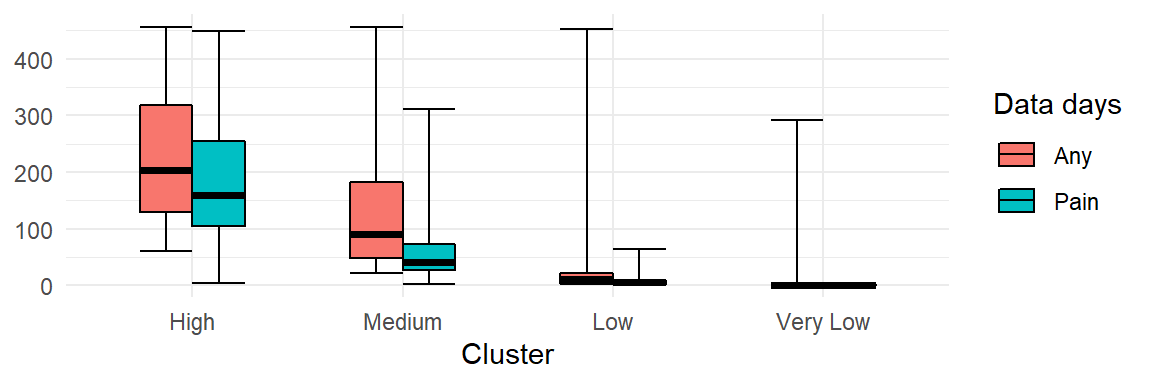
| Engagement | Cluster | Min | Q1 | Median | Q3 | Max |
|---|---|---|---|---|---|---|
| Days in study | High | 61 | 130 | 203 | 319 | 456 |
| Medium | 22 | 48 | 91 | 182 | ||
| Low | 2 | 4 | 10 | 21 | 452 | |
| Very Low | 1 | 1 | 1 | 1 | 291 | |
| No. days with pain data | High | 4 | 104 | 160 | 254 | 449 |
| Medium | 2 | 26 | 40 | 72 | 311 | |
| Low | 1 | 2 | 5 | 9 | 64 | |
| Very Low | 1 | 1 | 1 | 2 | ||
| % days with pain data | High | 73 | 82 | 89 | ||
| Medium | 38 | 52 | 67 | |||
| Low | 32 | 56 | 81 | |||
| Very Low | 100 | 100 | 100 |
Perhaps clearer representations of the same information:
Add styling
By default, a kableExtra-infused table (a kbl) is unformatted. You can make it look like the default knitr formatting again:
grouped_tbl <-
kable(my_tbl) %>%
pack_rows('Demographics', 2, 2) %>%
pack_rows('Health conditions', 3, 10)
grouped_tbl %>%
kable_styling()
| Variable | n | % |
|---|---|---|
| Number of participants | 2658 | 100.0 |
| Demographics | ||
| Female | 2210 | 83.1 |
| Health conditions | ||
| Rheumatoid arthritis | 506 | 19.0 |
| Osteoarthritis | 926 | 34.8 |
| Ankylosing spondylitis | 235 | 8.8 |
| Gout | 97 | 3.6 |
| Arthritis (unspecified) | 972 | 36.6 |
| Fibromyalgia | 665 | 25.0 |
| Chronic headache | 271 | 10.2 |
| Neuropathic pain | 371 | 14.0 |
Or better yet, pick from fancier themes and options:
grouped_tbl %>%
kable_paper(full_width = FALSE)
| Variable | n | % |
|---|---|---|
| Number of participants | 2658 | 100.0 |
| Demographics | ||
| Female | 2210 | 83.1 |
| Health conditions | ||
| Rheumatoid arthritis | 506 | 19.0 |
| Osteoarthritis | 926 | 34.8 |
| Ankylosing spondylitis | 235 | 8.8 |
| Gout | 97 | 3.6 |
| Arthritis (unspecified) | 972 | 36.6 |
| Fibromyalgia | 665 | 25.0 |
| Chronic headache | 271 | 10.2 |
| Neuropathic pain | 371 | 14.0 |
grouped_tbl %>%
kable_paper('striped', full_width = FALSE)
| Variable | n | % |
|---|---|---|
| Number of participants | 2658 | 100.0 |
| Demographics | ||
| Female | 2210 | 83.1 |
| Health conditions | ||
| Rheumatoid arthritis | 506 | 19.0 |
| Osteoarthritis | 926 | 34.8 |
| Ankylosing spondylitis | 235 | 8.8 |
| Gout | 97 | 3.6 |
| Arthritis (unspecified) | 972 | 36.6 |
| Fibromyalgia | 665 | 25.0 |
| Chronic headache | 271 | 10.2 |
| Neuropathic pain | 371 | 14.0 |
grouped_tbl %>%
kable_classic('striped', full_width = FALSE)
| Variable | n | % |
|---|---|---|
| Number of participants | 2658 | 100.0 |
| Demographics | ||
| Female | 2210 | 83.1 |
| Health conditions | ||
| Rheumatoid arthritis | 506 | 19.0 |
| Osteoarthritis | 926 | 34.8 |
| Ankylosing spondylitis | 235 | 8.8 |
| Gout | 97 | 3.6 |
| Arthritis (unspecified) | 972 | 36.6 |
| Fibromyalgia | 665 | 25.0 |
| Chronic headache | 271 | 10.2 |
| Neuropathic pain | 371 | 14.0 |
grouped_tbl %>%
kable_minimal(full_width = FALSE)
| Variable | n | % |
|---|---|---|
| Number of participants | 2658 | 100.0 |
| Demographics | ||
| Female | 2210 | 83.1 |
| Health conditions | ||
| Rheumatoid arthritis | 506 | 19.0 |
| Osteoarthritis | 926 | 34.8 |
| Ankylosing spondylitis | 235 | 8.8 |
| Gout | 97 | 3.6 |
| Arthritis (unspecified) | 972 | 36.6 |
| Fibromyalgia | 665 | 25.0 |
| Chronic headache | 271 | 10.2 |
| Neuropathic pain | 371 | 14.0 |
grouped_tbl %>%
kable_material('hover')
| Variable | n | % |
|---|---|---|
| Number of participants | 2658 | 100.0 |
| Demographics | ||
| Female | 2210 | 83.1 |
| Health conditions | ||
| Rheumatoid arthritis | 506 | 19.0 |
| Osteoarthritis | 926 | 34.8 |
| Ankylosing spondylitis | 235 | 8.8 |
| Gout | 97 | 3.6 |
| Arthritis (unspecified) | 972 | 36.6 |
| Fibromyalgia | 665 | 25.0 |
| Chronic headache | 271 | 10.2 |
| Neuropathic pain | 371 | 14.0 |
Worried about losing formatting? You can automatically save the resulting table as an image with kableExtra::as_image().
(However, this is not recommended because bitmap screenshots are neither scalable nor searchable and are inaccessible for people with disabilities.)
grouped_tbl %>%
kable_paper('striped', full_width = T) %>%
as_image(file = 'table.png', zoom = 3)

Check the documentation for kableExtra to see what else is possible. Results in other formats (such as PDF) may vary.
Further reading
\(\LaTeX\) package
booktabs:“It is not going too far to say that if you cannot create a table using the commands in this package, you should redesign it.”
ROpenSci did a feature comparison of the various table making packages back in 2017.