Hammersmith Imanet Data File Formats
Marie-Claude Asselin, So-Jin Holohan, Rainer Hinz, Federico Türkheimer
Hammersmith Imanet, Ltd.
Cyclotron Building
Hammersmith Hospital
Du Cane Road
LONDON
W12 0NN
Great BritainTel : (+44) 020 8383 3162
Fax : (+44) 020 8383 2029
WWW: http://www.hammersmithimanet.com
Table of contents
SIFs were introduced at Hammersmith Imanet for several reasons:
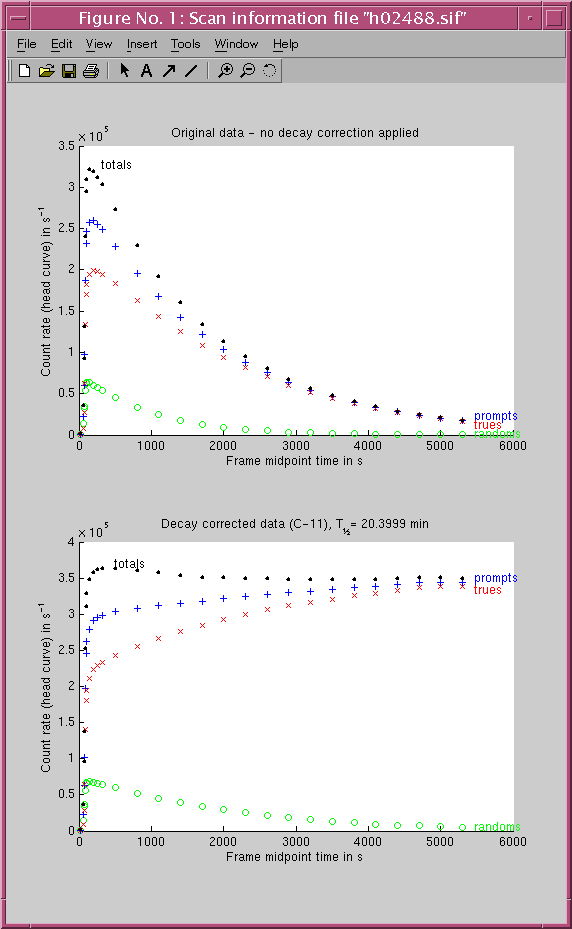
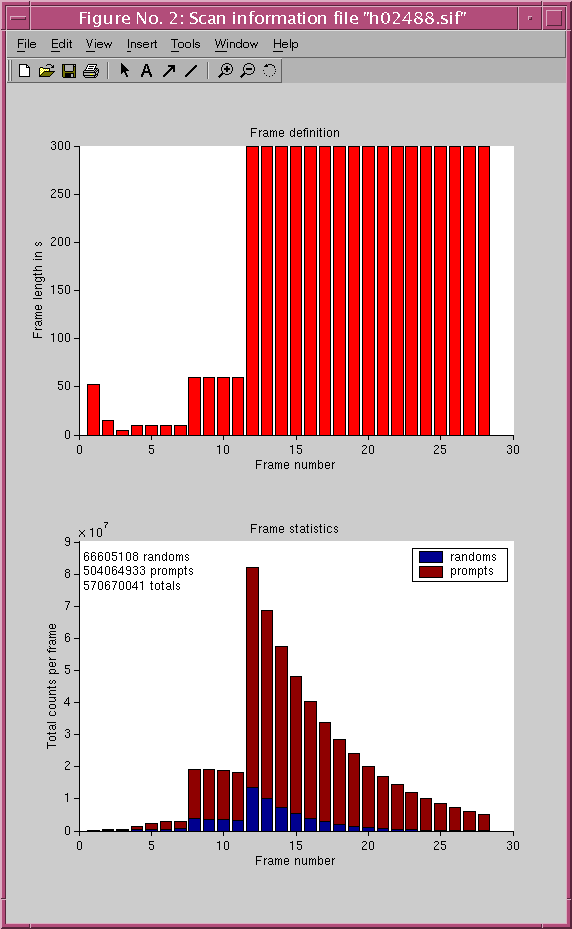
The contents of an example SIF is:
The first line is the header line. It should contain 7 fields separated by white spaces:31/1/2002 10:31:01 28 4 2 H02488 C-11
0 52 50617 46859
52 67 328520 206714
67 72 300377 162408
72 82 969573 350407
82 92 1874715 534353
92 102 2324417 618824
102 112 2463107 635273
112 172 15469060 3790914
172 232 15559987 3619122
232 292 15285747 3427281
292 352 14924555 3242744
352 652 68519120 13579865
652 952 58725752 9978332
952 1252 50185206 7300851
1252 1552 42741351 5303428
1552 1852 36406423 3868724
1852 2152 31060365 2812150
2152 2452 26476901 2039700
2452 2752 22567112 1480096
2752 3052 19154336 1066086
3052 3352 16272128 769421
3352 3652 13823059 553101
3652 3952 11744233 397199
3952 4252 9950919 285026
4252 4552 8457570 204345
4552 4852 7214108 149224
4852 5152 6084376 106504
5152 5452 5131299 76157


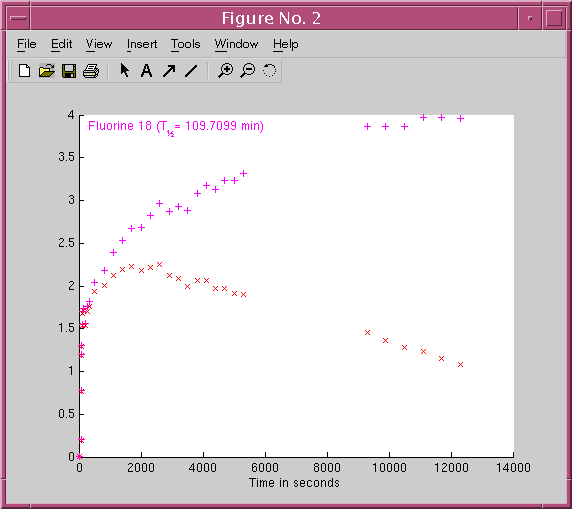
The first line is a header line. It contains the number of data points (lines) to read from the file after the header line. Anything written in the IDWC file after the number of lines specified in the header line should betreated as comments.34
29 0.00194262 2.5 1
65.5 0.197498 0.454485 1
75.5 0.771216 0.0294125 1
83 1.18433 0.0395171 1
93 1.29603 0.0350197 1
103 1.68379 0.032885 1
113 1.53591 0.0316392 1
148 1.71107 0.178779 1
208 1.53267 0.169358 1
268 1.70665 0.163331 1
328 1.76227 0.160366 1
508 1.9327 0.776158 1
808 2.00229 0.754066 1
1108 2.1239 0.735039 1
1408 2.18738 0.726569 1
1708 2.23053 0.720411 1
2008 2.17584 0.718495 1
2308 2.21135 0.723823 1
2608 2.25195 0.731343 1
2908 2.11765 0.741338 1
3208 2.0934 0.75195 1
3508 1.99675 0.765449 1
3808 2.06259 0.778843 1
4108 2.06017 0.795686 1
4408 1.96564 0.812938 1
4708 1.97341 0.829677 1
5008 1.90689 0.851037 1
5308 1.89793 0.869706 1
9300 1.45231 2.42591 1
9900 1.36335 2.58958 1
10500 1.28093 2.75193 1
11100 1.2335 2.92809 1
11700 1.15745 3.11498 1
12300 1.08516 3.3122 1
The first line is a header line. It contains the number of data points (lines) to read from the file after the header line. Anything written in the IF file after the number of lines specified in the header line should be treated as comments.5463
0.167969 0.00537605 0.00757944
1.27344 0.00537482 0.00757944
2.38281 -0.0372675 -0.0525658
3.48828 -0.0372589 -0.0525658
4.59375 -0.0372503 -0.0525658
5.69922 0.0479819 0.0677253
6.80469 -0.0372332 -0.0525658
7.91406 -0.0372245 -0.0525658
9.01953 0.00536615 0.00757944
10.125 0.00536491 0.00757944
: : :
: : :
: : :
: : :
5547 0.0142117 0.0543905
5548 0.0142033 0.0543588
5549 0.014195 0.0543272
5550 0.0141867 0.0542955
5551 0.0141783 0.0542638
5552 0.01417 0.0542322