Research Interests
The
biological physics group aims to apply techniques and ideas from
physics to biological problems. The group was established in 2001
and is currently developing teaching and research programmes. One of
the groupís strategic plans over the next few years is to expand
its size to a critical mass. Current research activities in the
group are described under the following topics
Our
work has centred on studying molecular structure and dynamics at wet
interfaces under conditions mimicking biological and biomedical
applications. We are well established in applying leading physical
techniques to access direct information at molecular and cellular
levels from various biointerfacial processes. The highlight of our
recent work has been to apply spectroscopic ellipsometry (SE) and
neutron reflection to reveal molecular features underlying surface
biocompatibility, a topic highly relevant to tissue engineering,
controlled local drug and gene delivery and medical implant
deployment.
We
often conduct these research activities in close collaboration with
life scientists and medical experts. Within the Biological Physics
Group, we place a strong emphasis to seek integrated approaches
between theoreticians, computational experts and experimentalists.
Tissue
Engineering
Study of interactions between biomaterial and vascular cells is
highly relevant to the control of cell seeding and tissue growth. In
this project, we emphasise the understanding of the mediation of
representative ECM proteins on the attachment, spreading and growth of
vascular cells. In parallel, the up and down regulations of key
proteins excreted by cells are also monitored. Using well fabricated
planar surfaces and polymeric films as model biomaterial, the
different biointerfaces are interrogated by a combined approach of
biochemical and biophysical techniques. The information so obtained is
highly useful towards future model development and growth of 3D tissue
constructs.
Fig.
1 shows how loading of a GFP labelled DNA strand varies with the
charge density in biocompatible polymer matrix (along X-axis) and film
thickness (along Y-axis).
400
Ň
2000
Ň 
4000
Ň

Biomaterials
Development
We utilise our strong expertise in polymer and surfactant
research to design short peptide sequences as biomaterials. There are
some 20 natural amino acids that are polar (hydrophilic), non-polar
(hydrophobic) and charged (positive and negative). They offer almost
endless choices for novel peptide surfactant design and synthesis.
Neutron reflection and small angle neutron scattering (SANS) are well
suited for revealing the nano-structures of these peptides assembled
at interfaces and their aggregates formed in bulk solution.
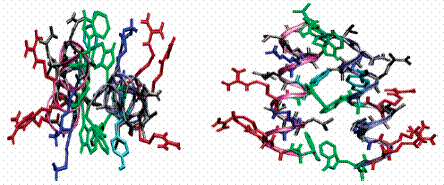
Fig.
2 Schematic of end (left) and side (right) views of a pair of 15-mer
peptides forming α-helical configuration via the strong
hydrophobic interdigitation of three tryptophans between them. The
α-helical backbone is illustrated as a ribbon. W groups are
labelled in green, Y in cyan, R in red, and K in blue. The α-helical
structure was revealed at the silicon oxide/water interface
(J. Am. Chem. Soc. 2004, 126, 8940).
In the last decade, advances in life sciences have gathered a vast amount of experimental information about biological systems at cellular and sub-cellular levels. Now we are facing two big challenges:
(1) how do we interpret, analyse the experimental information and relate it to the functions of life?
(2) Can we reconstruct biological systems from such detailed information?
There is a strong motivation to address these challenges. As a biological system is an integral system, within which components interact with each other. The interaction coordinates the simple behaviours of a biological system at cellular and sub-cellular levels into more complicated behaviours at tissue and organ levels. To understand the functions of a biological system, one has to synthesize the detailed, but isolated biological information obtained at cellular and sub-cellular systems into an interactive system at tissue and organ levels.
To tackle the two challenges requires multidisciplinary approaches. Recently, developments in non-linear science, modern physics of excitable medium, applied mathematics, together with availability of supercomputing power, have provided powerful tools to integrate detailed biological information into an interactive system. This forms a new exciting research area Ė reconstruction of virtual biological systems: from cell to organ.
Biomolecular structure and dynamics