
Dr. Andrew Gait
- Research Software Engineer
- IT Office 2
- Kilburn Building
- Research IT,
- University of Manchester,
- Oxford Road,
- Manchester
- M13 9PL
-
- Email: Andrew.Gait[at]manchester.ac.uk
Current work
I am now working as a Research Software Engineer as part of Research IT. I have two current projects: one relating to
image acquisition software for nailfold images used in the diagnosis of systemic sclerosis, and the other working on imaging
software to help with guidance for clinicians performing tracheostomies.
Previous work
I was part of the software team for the SpiNNaker project, led by Prof. Steve Furber, in the APT group at the Department of Computer Science.
This involved day-to-day support of users of the software across various applications, mainly related to neuromorphic
modelling and the Human Brain Project / EBRAINS. The software stack can be found on GitHub, along with associated documentation (including local installation instructions). You
can try SpiNNaker out for yourself by getting an EBRAINS account and using Jupyter notebooks.
See our paper that describes the sPyNNaker API, version 4.0, and another which describes the execution stages and engine beneath this: SpiNNTools. You can also look at the SpiNNaker book.
SpiNNaker is not necessarily limited to neuromorphic modelling: see my poster from RSE 2018.
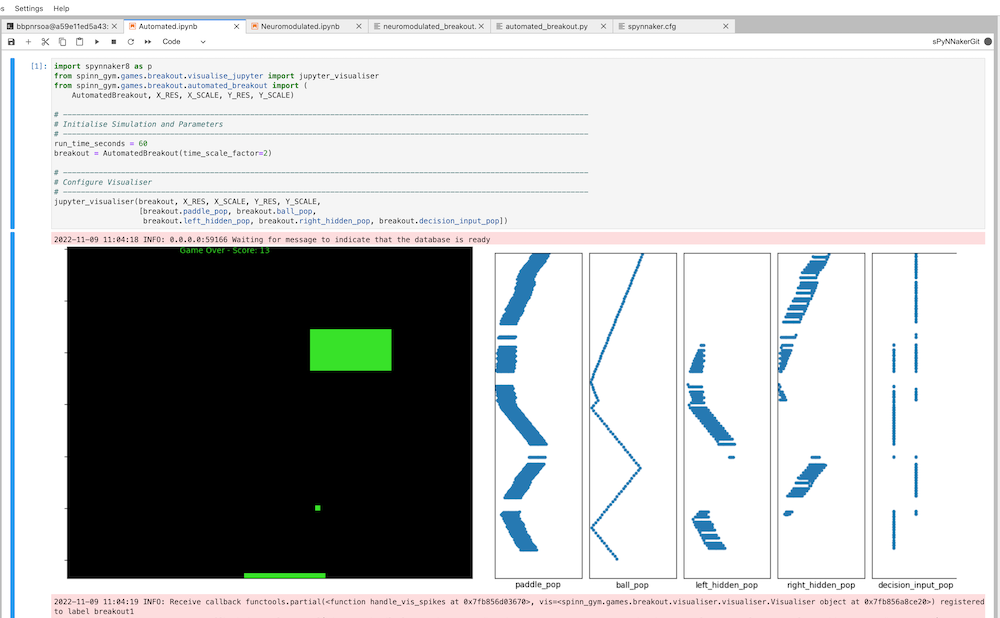
I worked at the Wolfson Molecular Imaging Centre on various software solutions to problems in image analysis. In particular, I designed a software tool to investigate parallel coordinates across multiple co-registered image sets.
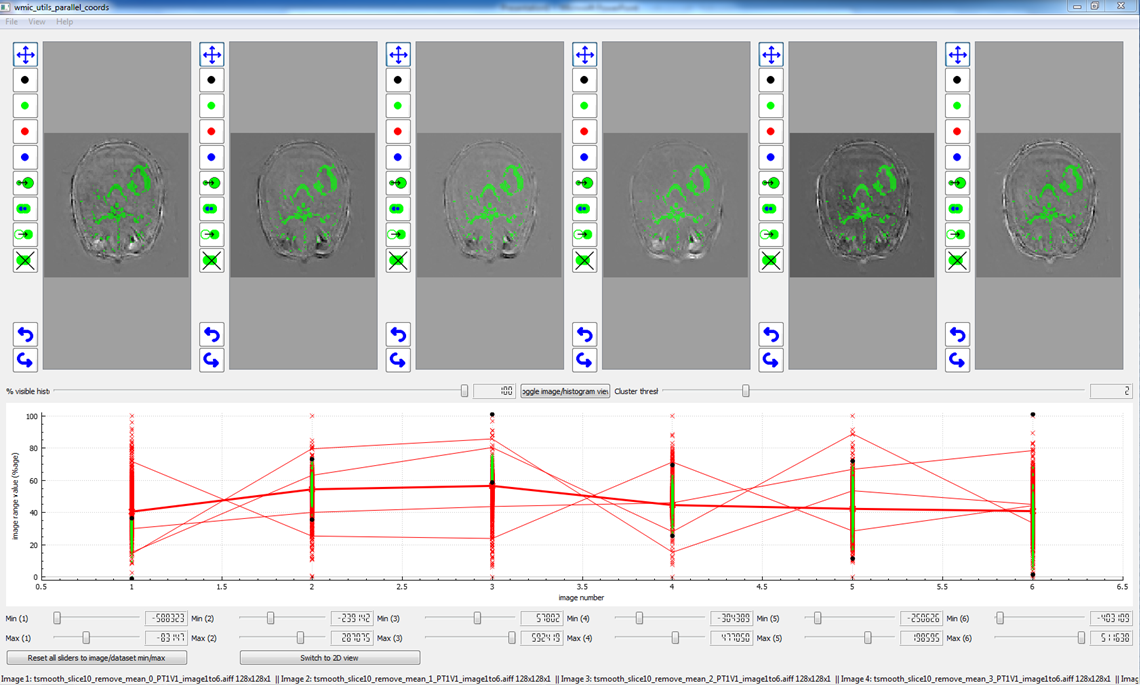
An example using the software tool for parallel coordinates with oxygen-enhanced MRI.
Previous to that (2009-2014), I worked on the Research into OsteoArthritis in Manchester (ROAM) project. I worked with Prof. Tim Cootes on image analysis of MRI scans of knees.
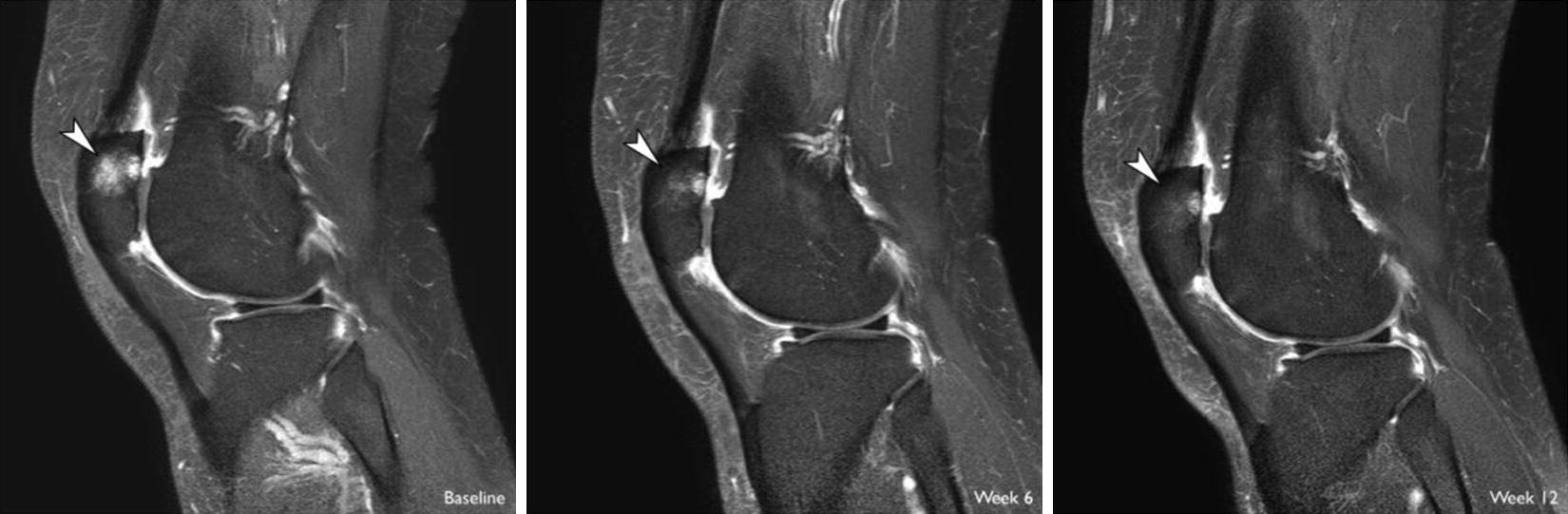
An image of the progression of a bone marrow lesion during treatment with a brace.
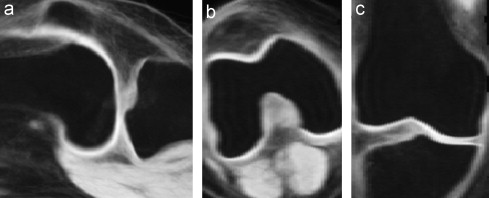
A mean image using generalised group-wise registration of the knees of 17 patients.
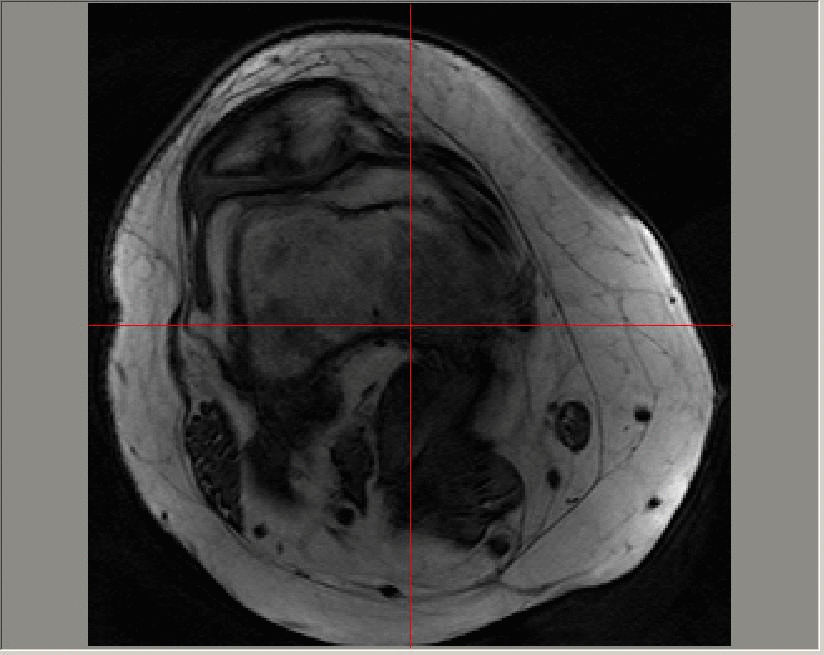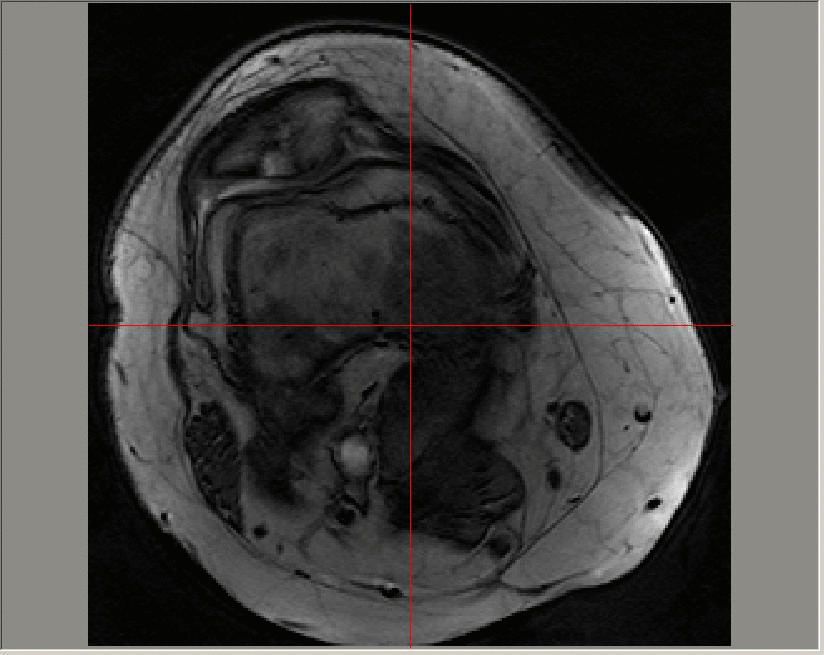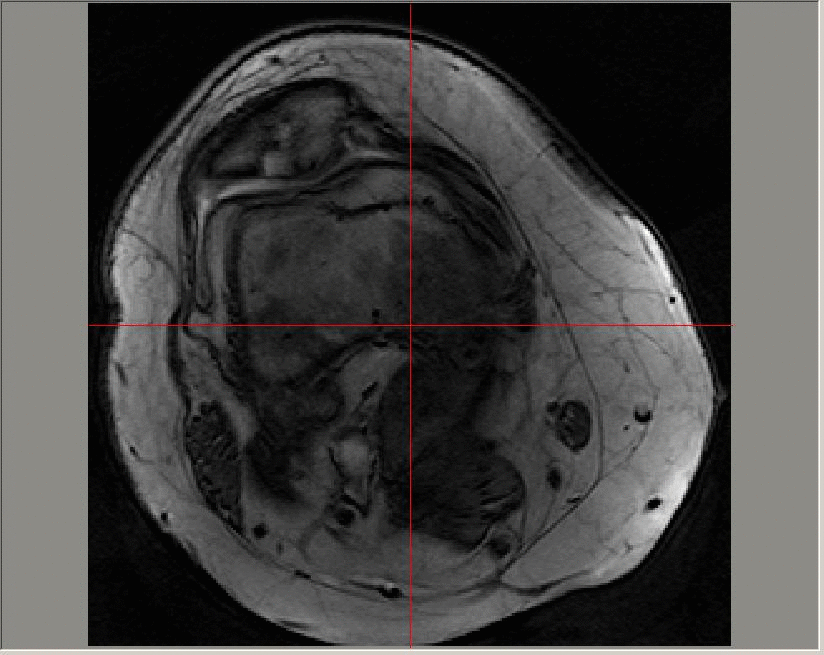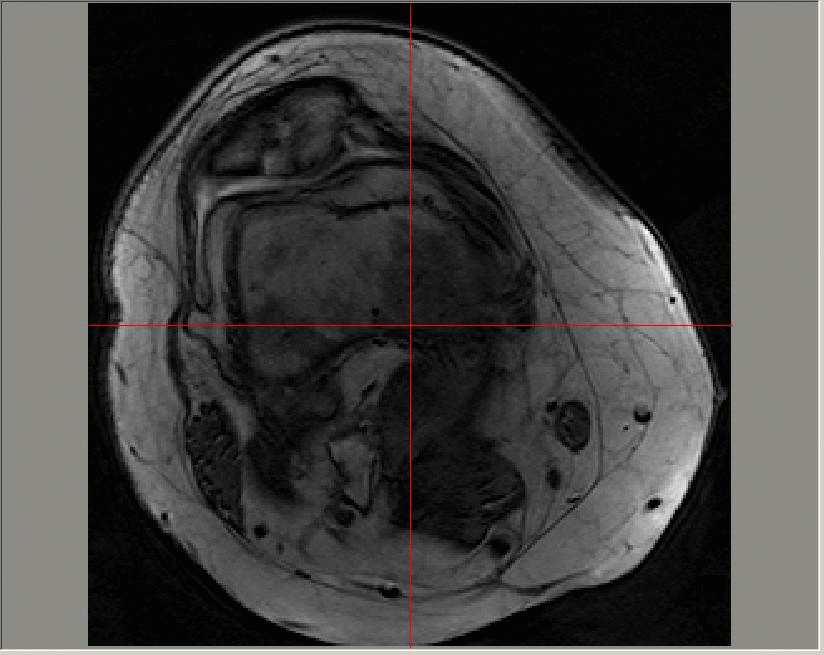
A Gadolinium-enhanced dynamic image sequence; images in this sequence are taken at a time resolution of roughly 23 seconds. The contrast agent is added at around 50-60 seconds and can be seen on the image first in the major artery, and later in the enhanced regions in the patellofemoral region towards the top of the image.
The uptake of the contrast agent can be modelled at each pixel on each slice of a 3D image using an exponential function (for example, using a Tofts model). The parameters that define the exponential function can then be averaged across diseased regions and longitudinal variation in the median values studied.

Previous to this (2007-2009), I worked on the oomph-lib project in the School of Mathematics at the University of Manchester, in particular on parallel domain decomposition techniques and associated communication requirements.
I completed my PhD at the School of Earth Sciences, University of Leeds, in May 2007.
Recent publications
For a complete list, see my ORCID profile.
- Perrett, A., Summerton. S., Gait. A. & Rhodes O., Online learning in SNNs with e-prop and Neuromorphic Hardware, NICE 2022: Neuro-Inspired Computational Elements Conference March 2022 Pages 32-39 10.1145/3517343.3517352
- Chiplunkar, C., Gautam, N., Mediratta, I., Gait, A., Thomas, S., Rowley, A., Serrano-Gotarredona, T. & Sen-Bhattacharya, B., A Reduced-Scale Cortical Network with Izhikevich's Neurons on SpiNNaker, 2021 International Joint Conference on Neural Networks (IJCNN), 2021, pp. 1-8, 10.1109/ijcnn52387.2021.9534244
- Rhodes, O., Peres, L., Rowley, A. G. D., Gait, A., Plana, L. A., Brenninkmeijer, C. & Furber, S.B., Real-time cortical simulation on neuromorphic hardware. Phil. Trans. R. Soc. A 378: 20190160, 23 December 2019, 10.1098/rsta.2019.0160
- Rowley, A. G. D., Brenninkmeijer, C., Davidson, S., Fellows, D., Gait, A., Lester, D. R., Mikaitis, M., Plana, L. A., Rhodes, O., Stokes, A. B. & Furber, S. B., SpiNNTools: The Execution Engine for the SpiNNaker Platform. Front. Neurosci., 26 March 2019, 10.3389/fnins.2019.00231
- Rhodes, O., Bogdan, P. A., Brenninkmeijer, C., Davidson, S., Fellows, D., Gait, A., Lester, D. R., Mikaitis, M., Plana, L. A., Rowley, A. G. D., Stokes, A. B. & Furber, S. B., sPyNNaker: A software package for running PyNN simulations on SpiNNaker. Front. Neurosci., 20 November 2018, 10.3389/fnins.2018.00816
- Perry, T., Gait, A., O'Neill, T. W., Parkes, M., Hodgson, R., Callaghan, M. J., Arden, N. K., Felson, D. T. & Cootes, T., Measurement of Synovial Tissue Volume in Knee Osteoarthritis using a Semi-automated MRI-based Quantitative Approach. Magn. Reson. Med. 2019;00:1-9, 15 Feb 2019, 10.1002/mrm.27633
Recent (first-author) conference presentations
- Gait, Andrew D. (2019). Running simulations on a massively-parallel low-power machine. Presentation at RSE 2019, University of Birmingham, UK.
- Gait, Andrew D., Yan, Juan, Hopkins, Michael, Rowley, Andrew G. D., Furber, Steve B. (2018). Implementing a multi-core algorithm on a massively parallel low-power system. Poster at RSE 2018, University of Birmingham, UK
- Gait, Andrew D., Hodgson, Richard, Cootes, Timothy F., Marjanovic, Elizabeth J., Parkes, Matthew J., O'Neill, Terrence W., Hutchinson, Charles E., Felson, David T. (2014). Late synovial enhancement detects effects of intra-articular steroids better than synovial volume [abstract]. Osteoarthritis and Cartilage Volume 22 Supplement, April 2014, pages S240-S241 DOI: 10.1016/j.joca.2014.02.465 - Poster at OARSI 2014, Paris, France
- Gait, Andrew D., Cootes, Timothy F., Marjanovic, Elizabeth J., Parkes, Matthew J., Hutchinson, Charles E., Felson, David T. (2012). Bone Marrow Lesions in Knees with Osteoarthritis: Can Parameters From Dynamic Contrast Enhancement Predict Change in Bone Marrow Lesion Volume or Knee Pain Change? [abstract]. Arthritis Rheum 2012;64 Suppl 10 :271 DOI: 10.1002/art.38006 - Poster at ACR 2012, Washington DC, USA
Past publications
- Gait, A.D., Hodgson, R., Parkes, M.J., Hutchinson, C.E., O'Neill, T.W., Maricar, N., Marjanovic, E.J., Cootes, T.F. & Felson, D.T., Synovial volume vs synovial measurements from dynamic contrast enhanced MRI as measures of response in osteoarthritis. Osteoarthritis & Cartilage, Vol. 24, Issue 8, Pages 1392-1398, August 2016, 10.1016/j.joca.2016.03.015.
- Kolawole Babalola, Andrew Gait, Timothy F. Cootes, A parts-and-geometry initialiser for 3D non-rigid registration using features derived from spin images, Neurocomputing, Volume 120, 23 November 2013, Pages 113-120, ISSN 0925-2312, 10.1016/j.neucom.2012.08.052.
- Felson, D., Parkes, M., Marjanovic, E., Callaghan, M., Gait, A., Cootes, T., Lunt, M., Oldham, J. & Hutchinson, C (2012). Bone marrow lesions in knee osteoarthritis change in 6 to 12 weeks. Osteoarthritis Cartilage, 20(12), 1514-1518. eScholarID:168316 | PMID:22944524 | DOI:10.1016/j.joca.2012.08.020
- Lowman, J. P., A. D. Gait, C. W. Gable, and H. Kukreja (2008), Plumes anchored by a high viscosity lower mantle in a 3D mantle convection model featuring dynamically evolving plates, Geophys. Res. Lett., 35, L19309, doi:10.1029/2008GL035342.
- Gait, A. D., J. P. Lowman, and C. W. Gable (2008), Time dependence in 3-D mantle convection models featuring evolving plates: Effect of lower mantle viscosity, J. Geophys. Res., 113, B08409, doi:10.1029/2007JB005538.
- Gait, A. D. and Lowman, J. P. (2007), Time-dependence in mantle convection models featuring dynamically evolving plates. Geophysical Journal International, 171: 463-477. doi: 10.1111/j.1365-246X.2007.03509.x.
- Gait, A. D., and J. P. Lowman (2007), Effect of lower mantle viscosity on the time-dependence of plate velocities in three-dimensional mantle convection models, Geophys. Res. Lett., 34, L21304, doi:10.1029/2007GL031396.






