3D Model Building and Matching
Research Associate: Kola Babalola
Principal Investigator: Tim Cootes
Project Period: October 2007 - April 2011
Contents
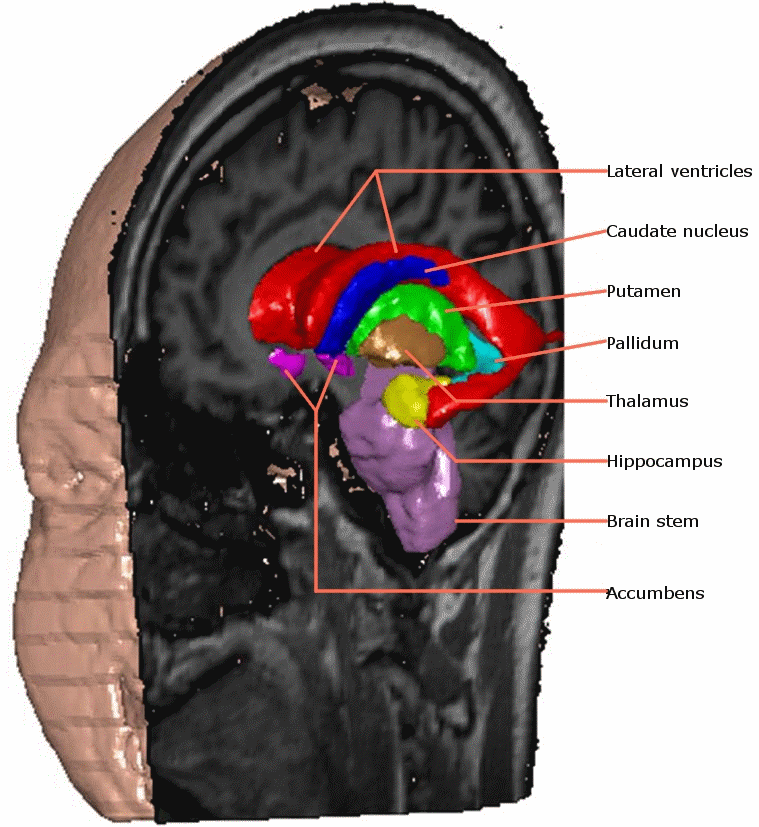
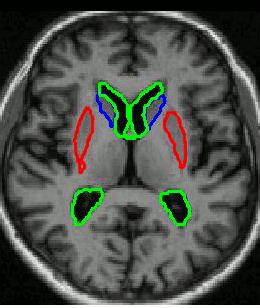
Research Associate: Kola Babalola
Principal Investigator: Tim Cootes
Project Period: October 2007 - April 2011


Image segmentation - delineating structure(s) of interest in an image, is a fundamental part of medical image processing. As imaging becomes more routinely used for diagnostic and therapeutic purposes, automating segmentation is necessary to increase throughput of processing, reduce the workload on staff involved and increase the quality of quantitative information derived from segmentation.
This EPSRC funded project addressed the automation of both the construction of 3D volumetric Active Appearance Models (AAMs) and their use to segment structures in medical images. Our AAM approach to segmentation offers the following advantages:
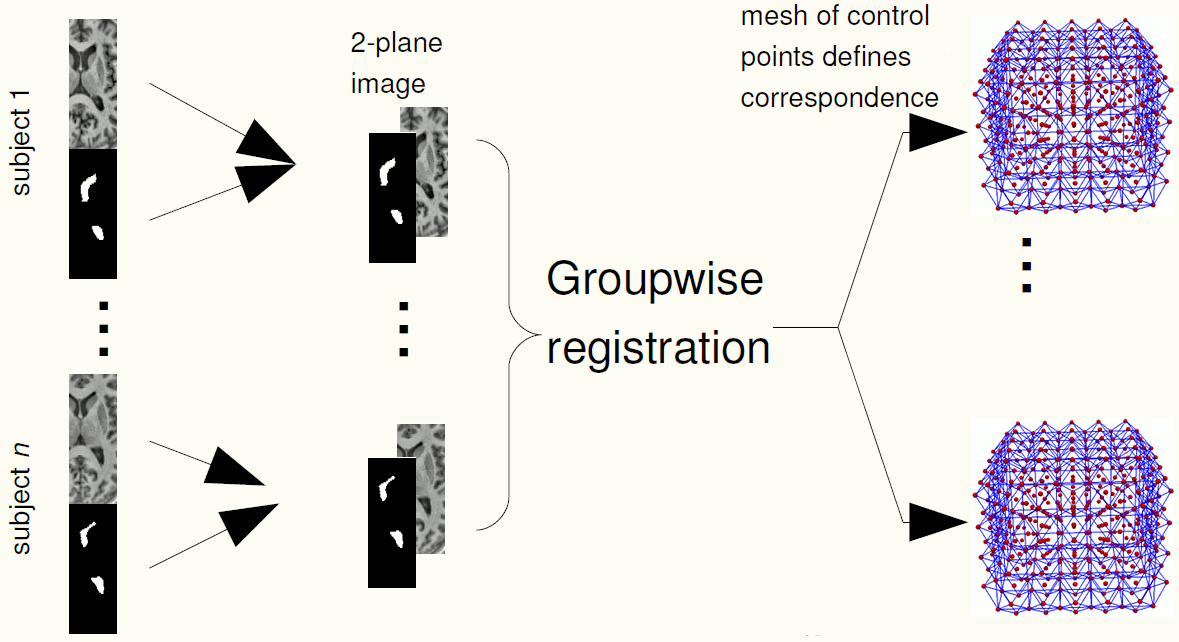
We use groupwise registration of 2-plane images (a labelled image and a gray level image) to establish correspondence across the training images. The AAM can be constructed using the correspondences.
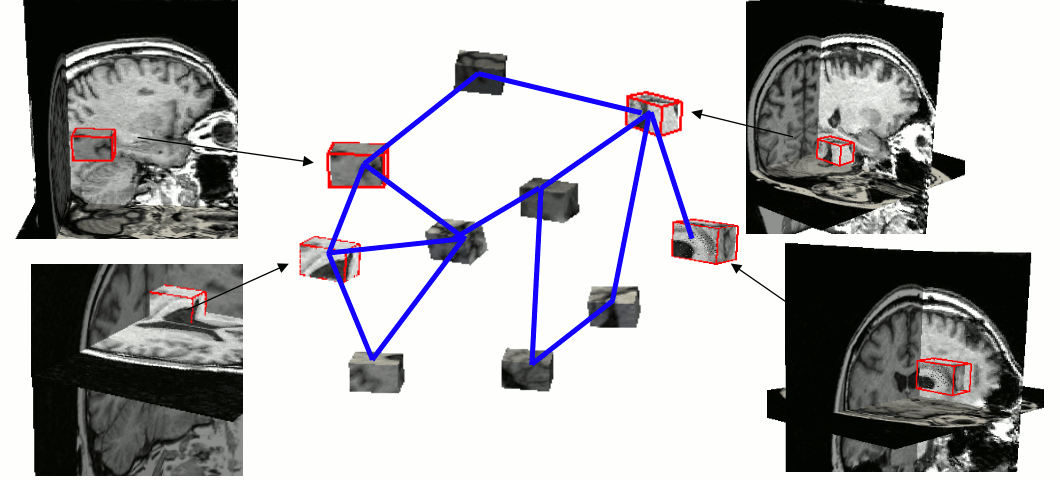
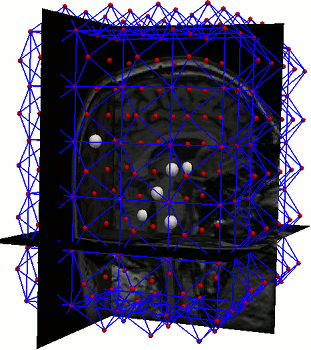
A parts-and-geometry model is a configuration of local feature detectors (the parts) and a set of relationships between their positions (the geometry). We automatically construct them by identifying local regions that can be reliably detected in most of the training set images
AAMs are a local optimisation method. We used parts-and-geometry models to automatically initialise the searches in 3D.
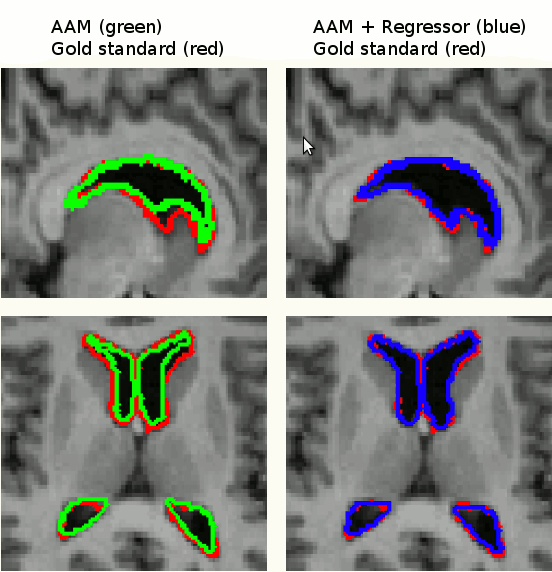
Segmentation by an AAM is based on finding model parameters to best fit a model to an image. There is therefore a residual difference between the approximated instance and the target image. We improved the results of segmentation by refining the search results using image data in a bottom-up approach using linear regressors and graph-cuts.
Executables for 3D AAMs to segment 10 subcortical structures are available here.
Back to contentsUsing parts and geometry models to initialise active appearance models for automated segmentation of 3D medical images. KO Babalola, TF Cootes, ISBI 2010: 1069-1072
Computing Accurate Correspondences Across Groups of Images. TF Cootes, CJ Twining, VS Petrovic, KO Babalola and CJ Taylor. IEEE Transactions on Pattern Analysis and Machine Intelligence 32(11): 1994-2005 (2010)
Brain serotonin transporter occupancy by oral sibutramine dosed to steady state: a PET study using 11C-DASB in healthy humans. PS Talbot, S Bradley, CP Clarke, KO Babalola, AW Philipp, G Brown, AW McMahon, JC Matthews, Neuropsychopharmacology: 35, 741-751 (2010)
AAM Segmentation of the Mandible and Brainstem. KO Babalola and TF Cootes. In Proceedings of the 3rd Workshop on 3D Segmentation in the Clinic: A Grand Challenge at the 12th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), September 2009
Serotonin transporter (SERT) occupancy by oral sibutramine in the living human brain using 11C-DASB PET P Talbot, S Bradley, C Clarke, K Babalola, A Philipp, G Brown, A McMahon, J Matthews. J NUCL MED MEETING ABSTRACTS vol 50(supplement 2) p 487, May 2009
3D Brain Segmentation Using Active Appearance Models and Local Regressors. KO Babalola, TF Cootes, CJ Twining, V Petrovic and CJ Taylor. In Proceedings of the 11th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), Lecture Notes in Computer Science, vol. 5241, pp. 410-408, September 2008
Automated segmentation of the caudate nuclei using Active Appearance Models KO Babalola, V Petrovic, TF Cootes, CJ Taylor, CJ Twining, TG Williams, and A Mills. In Proceedings of the First Workshop on 3D Segmentation in the Clinic: A Grand Challenge at the 10th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), pp. 57-64, November 2007
Back to contents